Hydroxyatrazine N-ethylaminohydrolase (AtzB): an amidohydrolase superfamily enzyme catalyzing deamination and dechlorination
- PMID: 17660279
- PMCID: PMC2045197
- DOI: 10.1128/JB.00630-07
Hydroxyatrazine N-ethylaminohydrolase (AtzB): an amidohydrolase superfamily enzyme catalyzing deamination and dechlorination
Abstract
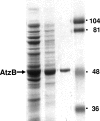
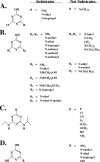
Hydroxyatrazine [2-(N-ethylamino)-4-hydroxy-6-(N-isopropylamino)-1,3,5-triazine] N-ethylaminohydrolase (AtzB) is the sole enzyme known to catalyze the hydrolytic conversion of hydroxyatrazine to N-isopropylammelide. AtzB, therefore, serves as the point of intersection of multiple s-triazine biodegradative pathways and is completely essential for microbial growth on s-triazine herbicides. Here, atzB was cloned from Pseudomonas sp. strain ADP and its product was purified to homogeneity and characterized. AtzB was found to be dimeric, with subunit and holoenzyme molecular masses of 52 kDa and 105 kDa, respectively. The k(cat) and K(m) of AtzB with hydroxyatrazine as a substrate were 3 s(-1) and 20 microM, respectively. Purified AtzB had a 1:1 zinc-to-subunit stoichiometry. Sequence analysis revealed that AtzB contained the conserved mononuclear amidohydrolase superfamily active-site residues His74, His76, His245, Glu248, His280, and Asp331. An intensive in vitro investigation into the substrate specificity of AtzB revealed that 20 of the 51 compounds tested were substrates for AtzB; this allowed for the identification of specific substrate structural features required for catalysis. Substrates required a monohydroxylated s-triazine ring with a minimum of one primary or secondary amine substituent and either a chloride or amine leaving group. AtzB catalyzed both deamination and dechlorination reactions with rates within a range of one order of magnitude. This differs from AtzA and TrzN, which do not catalyze deamination reactions, and AtzC, which is not known to catalyze dechlorination reactions.
Figures


Similar articles
-
AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes.J Bacteriol. 1998 Jan;180(1):152-8. doi: 10.1128/JB.180.1.152-158.1998. J Bacteriol. 1998. PMID: 9422605 Free PMC article.
-
The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway.Appl Environ Microbiol. 1997 Mar;63(3):916-23. doi: 10.1128/aem.63.3.916-923.1997. Appl Environ Microbiol. 1997. PMID: 9055410 Free PMC article.
-
Purification, substrate range, and metal center of AtzC: the N-isopropylammelide aminohydrolase involved in bacterial atrazine metabolism.J Bacteriol. 2002 Oct;184(19):5376-84. doi: 10.1128/JB.184.19.5376-5384.2002. J Bacteriol. 2002. PMID: 12218024 Free PMC article.
-
Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies.Appl Microbiol Biotechnol. 2002 Jan;58(1):39-45. doi: 10.1007/s00253-001-0862-y. Appl Microbiol Biotechnol. 2002. PMID: 11831474 Review.
-
Regulation of the atrazine-degradative genes in Pseudomonas sp. strain ADP.FEMS Microbiol Lett. 2010 Sep 1;310(1):1-8. doi: 10.1111/j.1574-6968.2010.01991.x. Epub 2010 Apr 15. FEMS Microbiol Lett. 2010. PMID: 20497226 Review.
Cited by
-
Microbial changes linked to the accelerated degradation of the herbicide atrazine in a range of temperate soils.Environ Sci Pollut Res Int. 2017 Mar;24(8):7359-7374. doi: 10.1007/s11356-017-8377-y. Epub 2017 Jan 20. Environ Sci Pollut Res Int. 2017. PMID: 28108915 Free PMC article.
-
A synthesis of the effects of pesticides on microbial persistence in aquatic ecosystems.Crit Rev Toxicol. 2015;45(10):813-36. doi: 10.3109/10408444.2015.1065471. Epub 2015 Nov 13. Crit Rev Toxicol. 2015. PMID: 26565685 Free PMC article. Review.
-
Plasmid localization and organization of melamine degradation genes in Rhodococcus sp. strain Mel.Appl Environ Microbiol. 2012 Mar;78(5):1397-403. doi: 10.1128/AEM.06468-11. Epub 2011 Dec 30. Appl Environ Microbiol. 2012. PMID: 22210223 Free PMC article.
-
Retracing the Rapid Evolution of an Herbicide-Degrading Enzyme by Protein Engineering.ACS Catal. 2023 Nov 17;13(23):15558-15571. doi: 10.1021/acscatal.3c04010. ACS Catal. 2023. PMID: 38567019 Free PMC article.
-
Thermostable cyanuric acid hydrolase from Moorella thermoacetica ATCC 39073.Appl Environ Microbiol. 2009 Nov;75(22):6986-91. doi: 10.1128/AEM.01605-09. Epub 2009 Sep 18. Appl Environ Microbiol. 2009. PMID: 19767460 Free PMC article.
References
-
- Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42.
-
- Baer, H. P., G. I. Drummond, and E. L. Duncan. 1966. Formation and deamination of adenosine by cardiac muscle enzymes. Mol. Pharmacol. 2:67-76. - PubMed
-
- Bar, H. P., and G. I. Drummond. 1966. On the mechanism of adenosine deaminase action. Biochem. Biophys. Res. Commun. 24:584-587. - PubMed
-
- Behki, R. M., and S. U. Khan. 1986. Degradation of atrazine by Pseudomonas: N-dealkylation and dehalogenation of atrazine and its metabolites. J. Agric. Food Chem. 34:746-749.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

