Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress
- PMID: 17660288
- PMCID: PMC2045231
- DOI: 10.1128/JB.00533-07
Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress
Erratum in
- J Bacteriol. 2007 Dec;186(24):9150
Abstract
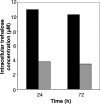
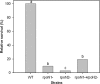
The growth and persistence of rhizobia and bradyrhizobia in soils are negatively impacted by drought conditions. In this study, we used genome-wide transcriptional analyses to obtain a comprehensive understanding of the response of Bradyrhizobium japonicum to drought. Desiccation of cells resulted in the differential expression of 15 to 20% of the 8,453 [corrected] B. japonicum open reading frames, with considerable differentiation between early (after 4 h) and late (after 24 and 72 h) expressed genes. While 225 genes were universally up-regulated at all three incubation times in response to desiccation, an additional 43 and 403 up-regulated genes were common to the 4/24- and 24/72-h incubation times, respectively. Desiccating conditions resulted in the significant induction (>2.0-fold) of the trehalose-6-phosphate synthetase (otsA), trehalose-6-phosphate phosphatase (otsB), and trehalose synthase (treS) genes, which encode two of the three trehalose synthesis pathways found in B. japonicum. Gene induction was correlated with an elevated intracellular concentration of trehalose and increased activity of trehalose-6-phosphate synthetase, collectively supporting the hypothesis that this disaccharide plays a prominent and important role in promoting desiccation tolerance in B. japonicum. Microarray data also indicated that sigma(54)- and sigma(24)-associated transcriptional regulators and genes encoding isocitrate lyase, oxidative stress responses, the synthesis and transport of exopolysaccharides, heat shock response proteins, enzymes for the modification and repair of nucleic acids, and the synthesis of pili and flagella are also involved in the response of B. japonicum to desiccation. Polyethylene glycol-generated osmotic stress induced significantly fewer genes than those transcriptionally activated by desiccation. However, 67 genes were commonly induced under both conditions. Taken together, these results suggest that B. japonicum directly responds to desiccation by adapting to changes imparted by reduced water activity, such as the synthesis of trehalose and polysaccharides and, secondarily, by the induction of a wide variety of proteins involved in protection of the cell membrane, repair of DNA damage, stability and integrity of proteins, and oxidative stress responses.
Figures



Similar articles
-
Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation.Appl Environ Microbiol. 2010 Feb;76(4):1071-81. doi: 10.1128/AEM.02483-09. Epub 2009 Dec 18. Appl Environ Microbiol. 2010. PMID: 20023090 Free PMC article.
-
Whole-genome expression profiling of Bradyrhizobium japonicum in response to hydrogen peroxide.Mol Plant Microbe Interact. 2011 Dec;24(12):1472-81. doi: 10.1094/MPMI-03-11-0072. Mol Plant Microbe Interact. 2011. PMID: 21864047
-
Effects of rehydration on physiological and transcriptional responses of a water-stressed rhizobium.J Microbiol. 2022 Jan;60(1):31-46. doi: 10.1007/s12275-022-1325-7. Epub 2021 Nov 26. J Microbiol. 2022. PMID: 34826097
-
Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression.Mol Microbiol. 1993 Apr;8(2):205-10. doi: 10.1111/j.1365-2958.1993.tb01564.x. Mol Microbiol. 1993. PMID: 8391102 Review.
-
Trehalose biosynthesis in response to abiotic stresses.J Integr Plant Biol. 2008 Oct;50(10):1223-9. doi: 10.1111/j.1744-7909.2008.00736.x. J Integr Plant Biol. 2008. PMID: 19017109 Review.
Cited by
-
Bradyrhizobium diazoefficiens Requires Chemical Chaperones To Cope with Osmotic Stress during Soybean Infection.mBio. 2021 Mar 30;12(2):e00390-21. doi: 10.1128/mBio.00390-21. mBio. 2021. PMID: 33785618 Free PMC article.
-
Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440.Antimicrob Agents Chemother. 2012 Feb;56(2):1001-9. doi: 10.1128/AAC.05398-11. Epub 2011 Dec 5. Antimicrob Agents Chemother. 2012. PMID: 22143519 Free PMC article.
-
Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1.Appl Environ Microbiol. 2008 May;74(9):2627-36. doi: 10.1128/AEM.02711-07. Epub 2008 Mar 7. Appl Environ Microbiol. 2008. PMID: 18326668 Free PMC article.
-
Enhanced tolerance to naphthalene and enhanced rhizoremediation performance for Pseudomonas putida KT2440 via the NAH7 catabolic plasmid.Appl Environ Microbiol. 2012 Aug;78(15):5104-10. doi: 10.1128/AEM.00619-12. Epub 2012 May 11. Appl Environ Microbiol. 2012. PMID: 22582075 Free PMC article.
-
Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures.Int J Mol Sci. 2016 May 27;17(6):815. doi: 10.3390/ijms17060815. Int J Mol Sci. 2016. PMID: 27240350 Free PMC article.
References
-
- Basaglia, M., S. Povolo, and S. Casella. 2007. Resuscitation of viable but not culturable Sinorhizobium meliloti 41 pRP4-luc: effects of oxygen and host plant. Curr. Microbiol. 54:167-174. - PubMed
-
- Benaroudj, N., D. H. Lee, and A. L. Goldberg. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261-24267. - PubMed
-
- Bergersen, F. J. 1961. The growth of rhizobia in synthetic media. Aust. J. Biol. Sci. 14:349-360.
-
- Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comp. Chem. 17:123-133.
-
- Boumahdi, M., P. Mary, and J. P. Hornez. 2001. Changes in fatty acid composition and degree of unsaturation of (brady)rhizobia as a response to phases of growth, reduced water activities and mild desiccation. Antonie Leeuwenhoek 79:73-79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

