Folding of a DNA hairpin loop structure in explicit solvent using replica-exchange molecular dynamics simulations
- PMID: 17660316
- PMCID: PMC2025651
- DOI: 10.1529/biophysj.107.108019
Folding of a DNA hairpin loop structure in explicit solvent using replica-exchange molecular dynamics simulations
Abstract
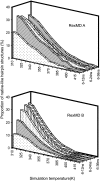
Hairpin loop structures are common motifs in folded nucleic acids. The 5'-GCGCAGC sequence in DNA forms a characteristic and stable trinucleotide hairpin loop flanked by a two basepair stem helix. To better understand the structure formation of this hairpin loop motif in atomic detail, we employed replica-exchange molecular dynamics (RexMD) simulations starting from a single-stranded DNA conformation. In two independent 36 ns RexMD simulations, conformations in very close agreement with the experimental hairpin structure were sampled as dominant conformations (lowest free energy state) during the final phase of the RexMDs ( approximately 35% at the lowest temperature replica). Simultaneous compaction and accumulation of folded structures were observed. Comparison of the GCA trinucleotides from early stages of the simulations with the folded topology indicated a variety of central loop conformations, but arrangements close to experiment that are sampled before the fully folded structure also appeared. Most of these intermediates included a stacking of the C(2) and G(3) bases, which was further stabilized by hydrogen bonding to the A(5) base and a strongly bound water molecule bridging the C(2) and A(5) in the DNA minor groove. The simulations suggest a folding mechanism where these intermediates can rapidly proceed toward the fully folded hairpin and emphasize the importance of loop and stem nucleotide interactions for hairpin folding. In one simulation, a loop motif with G(3) in syn conformation (dihedral flip at N-glycosidic bond) accumulated, resulting in a misfolded hairpin. Such conformations may correspond to long-lived trapped states that have been postulated to account for the folding kinetics of nucleic acid hairpins that are slower than expected for a semiflexible polymer of the same size.
Figures







Similar articles
-
Role of the closing base pair for d(GCA) hairpin stability: free energy analysis and folding simulations.Nucleic Acids Res. 2011 Oct;39(19):8271-80. doi: 10.1093/nar/gkr541. Epub 2011 Jun 30. Nucleic Acids Res. 2011. PMID: 21724608 Free PMC article.
-
Folding simulations of Trp-cage mini protein in explicit solvent using biasing potential replica-exchange molecular dynamics simulations.Proteins. 2009 Aug 1;76(2):448-60. doi: 10.1002/prot.22359. Proteins. 2009. PMID: 19173315
-
RNA simulations: probing hairpin unfolding and the dynamics of a GNRA tetraloop.J Mol Biol. 2002 Apr 5;317(4):493-506. doi: 10.1006/jmbi.2002.5447. J Mol Biol. 2002. PMID: 11955005
-
High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations.Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):16820-5. doi: 10.1073/pnas.1309392110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043821 Free PMC article. Review.
-
The RNA 3D Motif Atlas: Computational methods for extraction, organization and evaluation of RNA motifs.Methods. 2016 Jul 1;103:99-119. doi: 10.1016/j.ymeth.2016.04.025. Epub 2016 Apr 25. Methods. 2016. PMID: 27125735 Free PMC article. Review.
Cited by
-
Multimodal imaging and genetic characteristics of Chinese patients with USH2A-associated nonsyndromic retinitis pigmentosa.Mol Genet Genomic Med. 2020 Nov;8(11):e1479. doi: 10.1002/mgg3.1479. Epub 2020 Sep 6. Mol Genet Genomic Med. 2020. PMID: 32893482 Free PMC article.
-
Influence of Environmental Parameters on the Stability of the DNA Molecule.Entropy (Basel). 2021 Oct 31;23(11):1446. doi: 10.3390/e23111446. Entropy (Basel). 2021. PMID: 34828144 Free PMC article.
-
A kinetic zipper model with intrachain interactions applied to nucleic acid hairpin folding kinetics.Biophys J. 2012 Jan 4;102(1):101-11. doi: 10.1016/j.bpj.2011.11.4017. Epub 2012 Jan 3. Biophys J. 2012. PMID: 22225803 Free PMC article.
-
Role of the closing base pair for d(GCA) hairpin stability: free energy analysis and folding simulations.Nucleic Acids Res. 2011 Oct;39(19):8271-80. doi: 10.1093/nar/gkr541. Epub 2011 Jun 30. Nucleic Acids Res. 2011. PMID: 21724608 Free PMC article.
-
Structural Switch from Hairpin to Duplex/Antiparallel G-Quadruplex at Single-Nucleotide Polymorphism (SNP) Site of Human Apolipoprotein E (APOE) Gene Coding Region.ACS Omega. 2018 Mar 31;3(3):3173-3182. doi: 10.1021/acsomega.7b01654. Epub 2018 Mar 15. ACS Omega. 2018. PMID: 30023863 Free PMC article.
References
-
- Zhu, L., S. H. Chou, and B. R. Reid. 1996. Structure of a single cytidine hairpin loop formed by the DNA triplet GCA. Nat. Struct. Biol. 2:1012–1017. - PubMed
-
- Chou, S. H., L. Zhu, Z. Gao, J. W. Cheng, and B. R. Reid. 1996. Hairpin loops consisting of single adenine residues closed by sheared A:A or G:G pairs formed by DNA triplets AAA and GAG: solution structures of the d(GTACAAAGTAC) hairpin. J. Mol. Biol. 264:981–1001. - PubMed
-
- Chou, S. H., Y. Y. Tseng, and S. W. Wang. 1999. Stable sheared A:C pair in DNA hairpins. J. Mol. Biol. 287:301–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

