FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii
- PMID: 17660359
- PMCID: PMC2043389
- DOI: 10.1128/EC.00205-07
FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii
Abstract
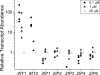
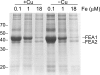
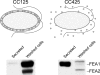
Previously, we had identified FOX1 and FTR1 as iron deficiency-inducible components of a high-affinity copper-dependent iron uptake pathway in Chlamydomonas. In this work, we survey the version 3.0 draft genome to identify a ferrireductase, FRE1, and two ZIP family proteins, IRT1 and IRT2, as candidate ferrous transporters based on their increased expression in iron-deficient versus iron-replete cells. In a parallel proteomic approach, we identified FEA1 and FEA2 as the major proteins secreted by iron-deficient Chlamydomonas reinhardtii. The recovery of FEA1 and FEA2 from the medium of Chlamydomonas strain CC425 cultures is strictly correlated with iron nutrition status, and the accumulation of the corresponding mRNAs parallels that of the Chlamydomonas FOX1 and FTR1 mRNAs, although the magnitude of regulation is more dramatic for the FEA genes. Like the FOX1 and FTR1 genes, the FEA genes do not respond to copper, zinc, or manganese deficiency. The 5' flanking untranscribed sequences from the FEA1, FTR1, and FOX1 genes confer iron deficiency-dependent expression of ARS2, suggesting that the iron assimilation pathway is under transcriptional control by iron nutrition. Genetic analysis suggests that the secreted proteins FEA1 and FEA2 facilitate high-affinity iron uptake, perhaps by concentrating iron in the vicinity of the cell. Homologues of FEA1 and FRE1 were identified previously as high-CO(2)-responsive genes, HCR1 and HCR2, in Chlorococcum littorale, suggesting that components of the iron assimilation pathway are responsive to carbon nutrition. These iron response components are placed in a proposed iron assimilation pathway for Chlamydomonas.
Figures


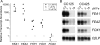


References
-
- Abboud, S., and D. J. Haile. 2000. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J. Biol. Chem. 275:19906-19912. - PubMed
-
- Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. - PubMed
-
- Askwith, C., and J. Kaplan. 1997. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272:401-405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

