High-density detection of restriction-site-associated DNA markers for rapid mapping of mutated loci in Neurospora
- PMID: 17660537
- PMCID: PMC2034621
- DOI: 10.1534/genetics.107.078147
High-density detection of restriction-site-associated DNA markers for rapid mapping of mutated loci in Neurospora
Abstract
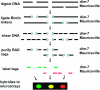
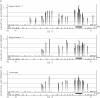
The wealth of sequence information available for Neurospora crassa and other fungi has greatly facilitated evolutionary and molecular analyses of this group. Although "reverse" genetics, in which genes are first identified by their sequence rather than by their mutant phenotypes, serves as a valuable new approach for elucidating biological processes, classical "forward" genetic analysis is still extremely useful. Unfortunately, mapping mutations and identifying the corresponding genes has typically been slow and laborious. To facilitate forward genetics in Neurospora, we have adapted microarray-based restriction-site-associated DNA (RAD) mapping for use with N. crassa oligonucleotide microarrays. This technique was used to simultaneously detect an unprecedented number of genomewide restriction site polymorphisms from two N. crassa strains: Mauriceville and Oak Ridge. Furthermore, RAD mapping was used to quickly map a previously unknown gene, defective in methylation-7 (dim-7).
Figures


Similar articles
-
The genome and genes of Neurospora crassa.Fungal Genet Biol. 1997 Jun;21(3):258-66. doi: 10.1006/fgbi.1997.0979. Fungal Genet Biol. 1997. PMID: 9290240 Review.
-
Genome-wide investigation of reproductive isolation in experimental lineages and natural species of Neurospora: identifying candidate regions by microarray-based genotyping and mapping.Evolution. 2010 Mar 1;64(3):694-709. doi: 10.1111/j.1558-5646.2009.00863.x. Epub 2009 Oct 5. Evolution. 2010. PMID: 19817850
-
A high-density single nucleotide polymorphism map for Neurospora crassa.Genetics. 2009 Feb;181(2):767-81. doi: 10.1534/genetics.108.089292. Epub 2008 Nov 17. Genetics. 2009. PMID: 19015548 Free PMC article.
-
Isolation of telomere DNA from Neurospora crassa.Mol Cell Biol. 1987 Sep;7(9):3168-77. doi: 10.1128/mcb.7.9.3168-3177.1987. Mol Cell Biol. 1987. PMID: 2890097 Free PMC article.
-
RIP: the evolutionary cost of genome defense.Trends Genet. 2004 Sep;20(9):417-23. doi: 10.1016/j.tig.2004.07.007. Trends Genet. 2004. PMID: 15313550 Review.
Cited by
-
Construction and application for QTL analysis of a Restriction Site Associated DNA (RAD) linkage map in barley.BMC Genomics. 2011 Jan 4;12:4. doi: 10.1186/1471-2164-12-4. BMC Genomics. 2011. PMID: 21205322 Free PMC article.
-
Rapid development of molecular markers by next-generation sequencing linked to a gene conferring phomopsis stem blight disease resistance for marker-assisted selection in lupin (Lupinus angustifolius L.) breeding.Theor Appl Genet. 2013 Feb;126(2):511-22. doi: 10.1007/s00122-012-1997-1. Epub 2012 Oct 20. Theor Appl Genet. 2013. PMID: 23086512
-
Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne.Theor Appl Genet. 2011 May;122(8):1467-80. doi: 10.1007/s00122-011-1546-3. Epub 2011 Feb 23. Theor Appl Genet. 2011. PMID: 21344184
-
Rapid mapping and identification of mutations in Caenorhabditis elegans by restriction site-associated DNA mapping and genomic interval pull-down sequencing.Genetics. 2011 Nov;189(3):767-78. doi: 10.1534/genetics.111.134031. Epub 2011 Sep 6. Genetics. 2011. PMID: 21900274 Free PMC article.
-
Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1.Eukaryot Cell. 2011 Jun;10(6):724-33. doi: 10.1128/EC.00016-11. Epub 2011 Apr 22. Eukaryot Cell. 2011. PMID: 21515825 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Bowring, F. J., and D. E. Catcheside, 1999. Recombinational landscape across a 650-kb contig on the right arm of linkage group V in Neurospora crassa. Curr. Genet. 36: 270–274. - PubMed
-
- Davis, R. H., and F. J. De Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17A: 47–143.
-
- Davis, R. H., and D. D. Perkins, 2002. Timeline: Neurospora—a model of model microbes. Nat. Rev. Genet. 3: 397–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

