Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe
- PMID: 17660542
- PMCID: PMC2013719
- DOI: 10.1534/genetics.107.077255
Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe
Abstract
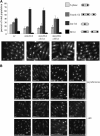
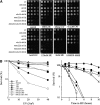
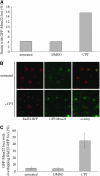
The faithful replication of the genome, coupled with the accurate repair of DNA damage, is essential for the maintenance of chromosomal integrity. The MMS22 gene of Saccharomyces cerevisiae plays an important but poorly understood role in preservation of genome integrity. Here we describe a novel gene in Schizosaccharomyces pombe that we propose is a highly diverged ortholog of MMS22. Fission yeast Mms22 functions in the recovery from replication-associated DNA damage. Loss of Mms22 results in the accumulation of spontaneous DNA damage in the S- and G2-phases of the cell cycle and elevated genomic instability. There are severe synthetic interactions involving mms22 and most of the homologous recombination proteins but not the structure-specific endonuclease Mus81-Eme1, which is required for survival of broken replication forks. Mms22 forms spontaneous nuclear foci and colocalizes with Rad22 in cells treated with camptothecin, suggesting that it has a direct role in repair of broken replication forks. Moreover, genetic interactions with components of the DNA replication fork suggest that Mms2 functions in the coordination of DNA synthesis following damage. We propose that Mms22 functions directly at the replication fork to maintain genomic integrity in a pathway involving Mus81-Eme1.
Figures





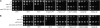
Similar articles
-
Mms1-Mms22 complex protects genome integrity in Schizosaccharomyces pombe.DNA Repair (Amst). 2009 Dec 3;8(12):1390-9. doi: 10.1016/j.dnarep.2009.09.008. Epub 2009 Oct 12. DNA Repair (Amst). 2009. PMID: 19819763 Free PMC article.
-
The novel gene mus7(+) is involved in the repair of replication-associated DNA damage in fission yeast.DNA Repair (Amst). 2007 Jun 1;6(6):770-80. doi: 10.1016/j.dnarep.2007.01.005. Epub 2007 Feb 20. DNA Repair (Amst). 2007. PMID: 17307401
-
Temporal separation of replication and recombination requires the intra-S checkpoint.J Cell Biol. 2005 Feb 14;168(4):537-44. doi: 10.1083/jcb.200410006. J Cell Biol. 2005. PMID: 15716375 Free PMC article.
-
Fission yeast mating-type switching: programmed damage and repair.DNA Repair (Amst). 2005 May 2;4(5):525-36. doi: 10.1016/j.dnarep.2004.11.004. Epub 2005 Jan 4. DNA Repair (Amst). 2005. PMID: 15811625 Review.
-
Linking the organization of DNA replication with genome maintenance.Curr Genet. 2019 Jun;65(3):677-683. doi: 10.1007/s00294-018-0923-8. Epub 2019 Jan 2. Curr Genet. 2019. PMID: 30600398 Review.
Cited by
-
Genetic Interaction Landscape Reveals Critical Requirements for Schizosaccharomyces pombe Brc1 in DNA Damage Response Mutants.G3 (Bethesda). 2015 Mar 19;5(5):953-62. doi: 10.1534/g3.115.017251. G3 (Bethesda). 2015. PMID: 25795664 Free PMC article.
-
The Role of Mms22p in DNA Damage Response in Candida albicans.G3 (Bethesda). 2015 Oct 4;5(12):2567-78. doi: 10.1534/g3.115.021840. G3 (Bethesda). 2015. PMID: 26438292 Free PMC article.
-
Regulation of rtt107 recruitment to stalled DNA replication forks by the cullin rtt101 and the rtt109 acetyltransferase.Mol Biol Cell. 2008 Jan;19(1):171-80. doi: 10.1091/mbc.e07-09-0961. Epub 2007 Oct 31. Mol Biol Cell. 2008. PMID: 17978089 Free PMC article.
-
The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination.Mol Cell. 2010 Nov 24;40(4):619-31. doi: 10.1016/j.molcel.2010.10.024. Epub 2010 Nov 4. Mol Cell. 2010. PMID: 21055983 Free PMC article.
-
Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method.Genome Res. 2013 Apr;23(4):705-15. doi: 10.1101/gr.146357.112. Epub 2012 Dec 17. Genome Res. 2013. PMID: 23249883 Free PMC article.
References
-
- Allshire, R. C., E. R. Nimmo, K. Ekwall, J.-P. Javerzat and G. Granston, 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233. - PubMed
-
- Araki, Y., Y. Kawasaki, H. Sasanuma, B. K. Tye and A. Sugino, 2003. Budding yeast mcm10/dna43 mutant requires a novel repair pathway for viability. Genes Cells 8: 465–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

