E(var)3-9 of Drosophila melanogaster encodes a zinc finger protein
- PMID: 17660546
- PMCID: PMC2013681
- DOI: 10.1534/genetics.107.076521
E(var)3-9 of Drosophila melanogaster encodes a zinc finger protein
Abstract
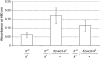
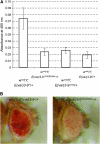
The importance of a gene's natural chromatin environment for its normal expression is poignantly illustrated when a change in chromosome position results in variable gene repression, such as is observed in position effect variegation (PEV) when the Drosophila melanogaster white (omega) gene is juxtaposed with heterochromatin. The Enhancer of variegation 3-9 [E(var)3-9] gene was one of over a hundred loci identified in screens for mutations that dominantly modify PEV. Haploinsufficiency for E(var)3-9 enhances omegam4 variegation, as would be expected from increased heterochromatin formation. To clarify the role of E(var)3-9 in chromosome structure, the gene has been cloned and its mutant alleles characterized. The involvement of E(var)3-9 in structure determination was supported by its reciprocal effects on euchromatic and heterochromatic PEV; E(var)3-9 mutations increased expression of a variegating heterochromatic gene in two tissue types. E(var)3-9 mutations also had a recessive phenotype, maternal effect lethality, which implicated E(var)3-9 function in an essential process during embryogenesis. Both phenotypes of E(var)3-9 mutations were consistent with its proposed function in promoting normal chromosome structure. The cloning of E(var)3-9 by classical genetic methods revealed that it encodes a protein with multiple zinc fingers, but otherwise novel sequence.
Figures




Similar articles
-
Suppression of heterochromatic gene variegation can be used to distinguish and characterize E(var) genes potentially important for chromosome structure in Drosophila melanogaster.Mol Genet Genomics. 2002 Feb;266(6):922-32. doi: 10.1007/s00438-001-0633-6. Epub 2002 Jan 25. Mol Genet Genomics. 2002. PMID: 11862486
-
Suppressors of position-effect variegation in Drosophila melanogaster affect expression of the heterochromatic gene light in the absence of a chromosome rearrangement.Genome. 1998 Aug;41(4):495-503. Genome. 1998. PMID: 9796098
-
Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a017780. doi: 10.1101/cshperspect.a017780. Cold Spring Harb Perspect Biol. 2013. PMID: 23906716 Free PMC article. Review.
-
A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster.Genetics. 2005 Apr;169(4):2165-77. doi: 10.1534/genetics.103.023341. Epub 2005 Jan 31. Genetics. 2005. PMID: 15687284 Free PMC article.
-
SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing.Genetica. 2003 Mar;117(2-3):149-58. doi: 10.1023/a:1022923508198. Genetica. 2003. PMID: 12723694 Review.
Cited by
-
The multi-AT-hook chromosomal protein of Drosophila melanogaster, D1, is dispensable for viability.Genetics. 2009 May;182(1):145-59. doi: 10.1534/genetics.109.101386. Epub 2009 Mar 16. Genetics. 2009. PMID: 19293138 Free PMC article.
-
High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model.Epigenetics Chromatin. 2009 Jan 29;2(1):1. doi: 10.1186/1756-8935-2-1. Epigenetics Chromatin. 2009. PMID: 19178722 Free PMC article.
-
Effete, a Drosophila chromatin-associated ubiquitin-conjugating enzyme that affects telomeric and heterochromatic position effect variegation.Genetics. 2013 Sep;195(1):147-58. doi: 10.1534/genetics.113.153320. Epub 2013 Jul 2. Genetics. 2013. PMID: 23821599 Free PMC article.
-
Heterochromatin formation in Drosophila requires genome-wide histone deacetylation in cleavage chromatin before mid-blastula transition in early embryogenesis.Chromosoma. 2020 Mar;129(1):83-98. doi: 10.1007/s00412-020-00732-x. Epub 2020 Jan 16. Chromosoma. 2020. PMID: 31950239 Free PMC article.
-
An accessible digital imaging workflow for multiplexed quantitative analysis of adult eye phenotypes in Drosophila melanogaster.bioRxiv [Preprint]. 2024 Aug 28:2024.01.26.577286. doi: 10.1101/2024.01.26.577286. bioRxiv. 2024. PMID: 39253516 Free PMC article. Preprint.
References
-
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas et al., 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
-
- Dorn, R., S. Heymann, R. Lindigkeit and G. Reuter, 1986. Suppressor mutations of position-effect variegation in Drosophila melanogaster affecting chromatin properties. Chromosoma 93: 398–403.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

