Triple mutants uncover three new genes required for social motility in Myxococcus xanthus
- PMID: 17660550
- PMCID: PMC2013723
- DOI: 10.1534/genetics.107.076182
Triple mutants uncover three new genes required for social motility in Myxococcus xanthus
Abstract
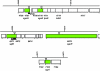
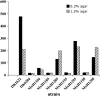
The bacterium Myxococcus xanthus glides over surfaces using two different locomotive mechanisms, called S (social) and A (adventurous) motility that enable cells to move both as groups and as individuals. Neither mechanism involves flagella. The functions of these two motors are coordinated by the activity of a small Ras-like protein, encoded by the mglA gene. The results of previous studies of a second-site suppressor of the mglA-8 missense mutation masK-815 indicate that MglA interacts with a protein tyrosine kinase, MasK, to control social motility. Sequence analysis of the sites of 12 independent insertions of the transposon magellan-4 that result in the loss of motility in an M. xanthus mglA-8 masK-815 double mutant shows that nine of these 12 insertions are in genes known to be required for S gliding motility. This result confirms that the masK-815 suppressor restores S but not A motility. Three of the 12 insertions define three new genes required for S motility and show that the attachment of heptose to the lipopolysaccharide inner core, an ortholog of the CheR methyltransferase, and a large protein with YD repeat motifs, are required for S motility. When these three insertions are backcrossed into an otherwise wild-type genetic background, their recombinants are found to have defects in S, but not, A motility. The spectrum of magellan-4 insertions that lead to the loss of S motility in the mglA-8 masK-815 double mutant background is different than that resulting from a previous mutant hunt starting with a different (A mutant) genetic background, suggesting that the number of genes required for S motility in M. xanthus is quite large.
Figures


Similar articles
-
Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner.Mol Microbiol. 2003 Jul;49(2):555-70. doi: 10.1046/j.1365-2958.2003.03582.x. Mol Microbiol. 2003. PMID: 12828649
-
MglA, a small GTPase, interacts with a tyrosine kinase to control type IV pili-mediated motility and development of Myxococcus xanthus.Mol Microbiol. 2002 Dec;46(5):1399-413. doi: 10.1046/j.1365-2958.2002.03258.x. Mol Microbiol. 2002. PMID: 12453225
-
Gliding motility in bacteria: insights from studies of Myxococcus xanthus.Microbiol Mol Biol Rev. 1999 Sep;63(3):621-41. doi: 10.1128/MMBR.63.3.621-641.1999. Microbiol Mol Biol Rev. 1999. PMID: 10477310 Free PMC article. Review.
-
MglC, a Paralog of Myxococcus xanthus GTPase-Activating Protein MglB, Plays a Divergent Role in Motility Regulation.J Bacteriol. 2015 Nov 16;198(3):510-20. doi: 10.1128/JB.00548-15. Print 2016 Feb 1. J Bacteriol. 2015. PMID: 26574508 Free PMC article.
-
Polarity of motility systems in Myxococcus xanthus.Curr Opin Microbiol. 2007 Dec;10(6):624-9. doi: 10.1016/j.mib.2007.09.012. Epub 2007 Nov 5. Curr Opin Microbiol. 2007. PMID: 17981496 Free PMC article. Review.
Cited by
-
Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links Type IVa pilus extrusion/retraction status to prey-independent growth signalling.PLoS One. 2013 Nov 5;8(11):e79759. doi: 10.1371/journal.pone.0079759. eCollection 2013. PLoS One. 2013. PMID: 24224002 Free PMC article.
-
Phylogenetic analysis of the teneurins: conserved features and premetazoan ancestry.Mol Biol Evol. 2012 Mar;29(3):1019-29. doi: 10.1093/molbev/msr271. Epub 2011 Oct 31. Mol Biol Evol. 2012. PMID: 22045996 Free PMC article.
-
The lifestyle switch protein Bd0108 of Bdellovibrio bacteriovorus is an intrinsically disordered protein.PLoS One. 2014 Dec 16;9(12):e115390. doi: 10.1371/journal.pone.0115390. eCollection 2014. PLoS One. 2014. PMID: 25514156 Free PMC article.
-
Members of the salivary gland surface protein (SGS) family are major immunogenic components of mosquito saliva.J Biol Chem. 2011 Nov 25;286(47):40824-34. doi: 10.1074/jbc.M111.280552. Epub 2011 Sep 29. J Biol Chem. 2011. PMID: 21965675 Free PMC article.
-
Cell division resets polarity and motility for the bacterium Myxococcus xanthus.J Bacteriol. 2014 Nov;196(22):3853-61. doi: 10.1128/JB.02095-14. Epub 2014 Aug 25. J Bacteriol. 2014. PMID: 25157084 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Bonner, P. J., Q. Xu, W. P. Black, Z. Li, Z. Yang and L. J. Shimkets 2005. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol. Microbiol. 57: 1499–1508. - PubMed
-
- Bowden, M. G., and H. B. Kaplan, 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30: 275–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

