Alternative splicing gives rise to different isoforms of the Neurospora crassa Tob55 protein that vary in their ability to insert beta-barrel proteins into the outer mitochondrial membrane
- PMID: 17660559
- PMCID: PMC2013688
- DOI: 10.1534/genetics.107.075051
Alternative splicing gives rise to different isoforms of the Neurospora crassa Tob55 protein that vary in their ability to insert beta-barrel proteins into the outer mitochondrial membrane
Abstract
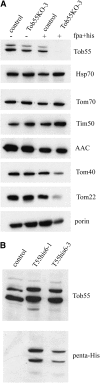
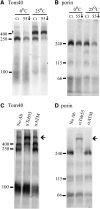
Tob55 is the major component of the TOB complex, which is found in the outer membrane of mitochondria. A sheltered knockout of the tob55 gene was developed in Neurospora crassa. When grown under conditions that reduce the levels of the Tob55 protein, the strain exhibited a reduced growth rate and mitochondria isolated from these cells were deficient in their ability to import beta-barrel proteins. Surprisingly, Western blots of wild-type mitochondrial proteins revealed two bands for Tob55 that differed by approximately 4 kDa in their apparent molecular masses. Sequence analysis of cDNAs revealed that the tob55 mRNA is alternatively spliced and encodes three isoforms of the protein, which are predicted to contain 521, 516, or 483 amino acid residues. Mass spectrometry of proteins isolated from purified outer membrane vesicles confirmed the existence of each isoform in mitochondria. Strains that expressed each isoform of the protein individually were constructed. When cells expressing only the longest form of the protein were grown at elevated temperature, their growth rate was reduced and mitochondria isolated from these cells were deficient in their ability to assembly beta-barrel proteins.
Figures






Similar articles
-
Analysis of mutations in Neurospora crassa ERMES components reveals specific functions related to β-barrel protein assembly and maintenance of mitochondrial morphology.PLoS One. 2013 Aug 5;8(8):e71837. doi: 10.1371/journal.pone.0071837. Print 2013. PLoS One. 2013. PMID: 23940790 Free PMC article.
-
The Neurospora crassa TOB complex: analysis of the topology and function of Tob38 and Tob37.PLoS One. 2011;6(9):e25650. doi: 10.1371/journal.pone.0025650. Epub 2011 Sep 28. PLoS One. 2011. PMID: 21980517 Free PMC article.
-
Structural requirements of Tom40 for assembly into preexisting TOM complexes of mitochondria.Mol Biol Cell. 2001 May;12(5):1189-98. doi: 10.1091/mbc.12.5.1189. Mol Biol Cell. 2001. PMID: 11359915 Free PMC article.
-
Biogenesis of beta-barrel membrane proteins of mitochondria.Trends Biochem Sci. 2005 Oct;30(10):575-82. doi: 10.1016/j.tibs.2005.08.009. Trends Biochem Sci. 2005. PMID: 16126389 Review.
-
Protein import into mitochondria of Neurospora crassa.Fungal Genet Biol. 2002 Jul;36(2):85-90. doi: 10.1016/S1087-1845(02)00018-X. Fungal Genet Biol. 2002. PMID: 12081461 Review.
Cited by
-
Transcriptome analysis of two isolates of the tomato pathogen Cladosporium fulvum, uncovers genome-wide patterns of alternative splicing during a host infection cycle.PLoS Pathog. 2024 Dec 18;20(12):e1012791. doi: 10.1371/journal.ppat.1012791. eCollection 2024 Dec. PLoS Pathog. 2024. PMID: 39693392 Free PMC article.
-
Analysis of mutations in Neurospora crassa ERMES components reveals specific functions related to β-barrel protein assembly and maintenance of mitochondrial morphology.PLoS One. 2013 Aug 5;8(8):e71837. doi: 10.1371/journal.pone.0071837. Print 2013. PLoS One. 2013. PMID: 23940790 Free PMC article.
-
Alternative function for the mitochondrial SAM complex in biogenesis of alpha-helical TOM proteins.J Cell Biol. 2007 Dec 3;179(5):881-93. doi: 10.1083/jcb.200706043. Epub 2007 Nov 26. J Cell Biol. 2007. PMID: 18039934 Free PMC article.
-
Evidence supporting the 19 β-strand model for Tom40 from cysteine scanning and protease site accessibility studies.J Biol Chem. 2014 Aug 1;289(31):21640-50. doi: 10.1074/jbc.M114.578765. Epub 2014 Jun 19. J Biol Chem. 2014. PMID: 24947507 Free PMC article.
-
Characterization of Single Gene Deletion Mutants Affecting Alternative Oxidase Production in Neurospora crassa: Role of the yvh1 Gene.Microorganisms. 2020 Aug 4;8(8):1186. doi: 10.3390/microorganisms8081186. Microorganisms. 2020. PMID: 32759834 Free PMC article.
References
-
- Ausubel, R. A., R. Brent, R. E. Kingston, D. D. Moore and J. G. Seidman, 1992. Current Protocols in Molecular Biology. Greene and Wiley Interscience, New York.
-
- Bos, M. P., and J. Tommassen, 2004. Biogenesis of the Gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7: 610–616. - PubMed
-
- Davis, R. H., and F. J. De Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17: 79–143.
-
- Dolezal, P., V. Likic, J. Tachezy and T. Lithgow, 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313: 314–318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

