Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1
- PMID: 17660562
- PMCID: PMC2013730
- DOI: 10.1534/genetics.107.074476
Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1
Abstract
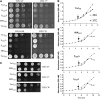
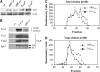
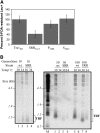
Tra1 is an essential component of the Saccharomyces cerevisiae SAGA and NuA4 complexes. Using targeted mutagenesis, we identified residues within its C-terminal phosphatidylinositol-3-kinase (PI3K) domain that are required for function. The phenotypes of tra1-P3408A, S3463A, and SRR3413-3415AAA included temperature sensitivity and reduced growth in media containing 6% ethanol or calcofluor white or depleted of phosphate. These alleles resulted in a twofold or greater change in expression of approximately 7% of yeast genes in rich media and reduced activation of PHO5 and ADH2 promoters. Tra1-SRR3413 associated with components of both the NuA4 and SAGA complexes and with the Gal4 transcriptional activation domain similar to wild-type protein. Tra1-SRR3413 was recruited to the PHO5 promoter in vivo but gave rise to decreased relative amounts of acetylated histone H3 and histone H4 at SAGA and NuA4 regulated promoters. Distinct from other components of these complexes, tra1-SRR3413 resulted in generation-dependent telomere shortening and synthetic slow growth in combination with deletions of a number of genes with roles in membrane-related processes. While the tra1 alleles have some phenotypic similarities with deletions of SAGA and NuA4 components, their distinct nature may arise from the simultaneous alteration of SAGA and NuA4 functions.
Figures




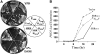
Similar articles
-
Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1.Curr Genet. 2010 Oct;56(5):447-65. doi: 10.1007/s00294-010-0313-3. Epub 2010 Jul 16. Curr Genet. 2010. PMID: 20635087 Free PMC article.
-
The Pseudokinase Domain of Saccharomyces cerevisiae Tra1 Is Required for Nuclear Localization and Incorporation into the SAGA and NuA4 Complexes.G3 (Bethesda). 2018 May 31;8(6):1943-1957. doi: 10.1534/g3.118.200288. G3 (Bethesda). 2018. PMID: 29626083 Free PMC article.
-
Systematic genetic array analysis links the Saccharomyces cerevisiae SAGA/SLIK and NuA4 component Tra1 to multiple cellular processes.BMC Genet. 2008 Jul 10;9:46. doi: 10.1186/1471-2156-9-46. BMC Genet. 2008. PMID: 18616809 Free PMC article.
-
Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes.Transcription. 2019 Feb;10(1):37-43. doi: 10.1080/21541264.2018.1530936. Epub 2018 Oct 30. Transcription. 2019. PMID: 30375921 Free PMC article. Review.
-
NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components.Biochem Cell Biol. 2009 Oct;87(5):799-815. doi: 10.1139/O09-062. Biochem Cell Biol. 2009. PMID: 19898529 Review.
Cited by
-
Tra1 controls the transcriptional landscape of the aging cell.G3 (Bethesda). 2023 Jan 12;13(1):jkac287. doi: 10.1093/g3journal/jkac287. G3 (Bethesda). 2023. PMID: 36315064 Free PMC article.
-
Genetic evidence links the ASTRA protein chaperone component Tti2 to the SAGA transcription factor Tra1.Genetics. 2012 Jul;191(3):765-80. doi: 10.1534/genetics.112.140459. Epub 2012 Apr 13. Genetics. 2012. PMID: 22505622 Free PMC article.
-
Loss of nonsense mediated decay suppresses mutations in Saccharomyces cerevisiae TRA1.BMC Genet. 2012 Mar 22;13:19. doi: 10.1186/1471-2156-13-19. BMC Genet. 2012. PMID: 22439631 Free PMC article.
-
Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1.Curr Genet. 2010 Oct;56(5):447-65. doi: 10.1007/s00294-010-0313-3. Epub 2010 Jul 16. Curr Genet. 2010. PMID: 20635087 Free PMC article.
-
The Pseudokinase Domain of Saccharomyces cerevisiae Tra1 Is Required for Nuclear Localization and Incorporation into the SAGA and NuA4 Complexes.G3 (Bethesda). 2018 May 31;8(6):1943-1957. doi: 10.1534/g3.118.200288. G3 (Bethesda). 2018. PMID: 29626083 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1998. Protocols in Molecular Biology. Greene/Wiley-Interscience, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

