DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes
- PMID: 17660745
- PMCID: PMC1952222
- DOI: 10.1038/sj.emboj.7601807
DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes
Abstract
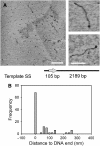
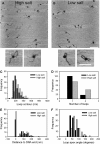
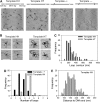
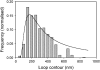
EcoP15I is a type III restriction enzyme that requires two recognition sites in a defined orientation separated by up to 3.5 kbp to efficiently cleave DNA. The mechanism through which site-bound EcoP15I enzymes communicate between the two sites is unclear. Here, we use atomic force microscopy to study EcoP15I-DNA pre-cleavage complexes. From the number and size distribution of loops formed, we conclude that the loops observed do not result from translocation, but are instead formed by a contact between site-bound EcoP15I and a nonspecific region of DNA. This conclusion is confirmed by a theoretical polymer model. It is further shown that translocation must play some role, because when translocation is blocked by a Lac repressor protein, DNA cleavage is similarly blocked. On the basis of these results, we present a model for restriction by type III restriction enzymes and highlight the similarities between this and other classes of restriction enzymes.
Figures


Similar articles
-
Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I.J Mol Biol. 2004 Aug 6;341(2):337-43. doi: 10.1016/j.jmb.2004.06.031. J Mol Biol. 2004. PMID: 15276827
-
Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping.Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12755-60. doi: 10.1073/pnas.0700483104. Epub 2007 Jul 23. Proc Natl Acad Sci U S A. 2007. PMID: 17646654 Free PMC article.
-
DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites.J Mol Biol. 2001 Sep 28;312(4):687-98. doi: 10.1006/jmbi.2001.4998. J Mol Biol. 2001. PMID: 11575924
-
Translocation, switching and gating: potential roles for ATP in long-range communication on DNA by Type III restriction endonucleases.Biochem Soc Trans. 2011 Apr;39(2):589-94. doi: 10.1042/BST0390589. Biochem Soc Trans. 2011. PMID: 21428945 Free PMC article. Review.
-
DNA translocation by type III restriction enzymes: a comparison of current models of their operation derived from ensemble and single-molecule measurements.Nucleic Acids Res. 2011 Jun;39(11):4525-31. doi: 10.1093/nar/gkq1285. Epub 2011 Feb 10. Nucleic Acids Res. 2011. PMID: 21310716 Free PMC article. Review.
Cited by
-
Direct visualization of G-quadruplexes in DNA using atomic force microscopy.Nucleic Acids Res. 2009 Oct;37(18):6269-75. doi: 10.1093/nar/gkp679. Epub 2009 Aug 20. Nucleic Acids Res. 2009. PMID: 19696072 Free PMC article.
-
Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems.DNA Res. 2010 Dec;17(6):325-42. doi: 10.1093/dnares/dsq027. Epub 2010 Nov 8. DNA Res. 2010. PMID: 21059708 Free PMC article. Review.
-
Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I.Nat Commun. 2015 Jun 12;6:7363. doi: 10.1038/ncomms8363. Nat Commun. 2015. PMID: 26067164 Free PMC article.
-
Single-site DNA cleavage by Type III restriction endonuclease requires a site-bound enzyme and a trans-acting enzyme that are ATPase-activated.Nucleic Acids Res. 2018 Jul 6;46(12):6229-6237. doi: 10.1093/nar/gky344. Nucleic Acids Res. 2018. PMID: 29846668 Free PMC article.
-
MmeI: a minimal Type II restriction-modification system that only modifies one DNA strand for host protection.Nucleic Acids Res. 2008 Nov;36(20):6558-70. doi: 10.1093/nar/gkn711. Epub 2008 Oct 17. Nucleic Acids Res. 2008. PMID: 18931376 Free PMC article.
References
-
- Bianco PR, Hurley EM (2005) The type I restriction endonuclease EcoR1241, couples ATP hydrolysis to bidirectional DNA translocation. J Mol Biol 352: 837–859 - PubMed
-
- Bist P, Sistla S, Krishnamurthy V, Acharya A, Chandrakala B, Rao DN (2001) S-adenosyl-L-methionine is required for DNA cleavage by type III restriction enzymes. J Mol Biol 310: 93–109 - PubMed
-
- Bourniquel AA, Bickle TA (2002) Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84: 1047–1059 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

