DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes
- PMID: 17660745
- PMCID: PMC1952222
- DOI: 10.1038/sj.emboj.7601807
DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes
Abstract
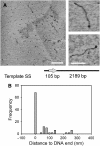
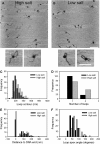
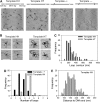
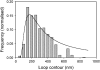
EcoP15I is a type III restriction enzyme that requires two recognition sites in a defined orientation separated by up to 3.5 kbp to efficiently cleave DNA. The mechanism through which site-bound EcoP15I enzymes communicate between the two sites is unclear. Here, we use atomic force microscopy to study EcoP15I-DNA pre-cleavage complexes. From the number and size distribution of loops formed, we conclude that the loops observed do not result from translocation, but are instead formed by a contact between site-bound EcoP15I and a nonspecific region of DNA. This conclusion is confirmed by a theoretical polymer model. It is further shown that translocation must play some role, because when translocation is blocked by a Lac repressor protein, DNA cleavage is similarly blocked. On the basis of these results, we present a model for restriction by type III restriction enzymes and highlight the similarities between this and other classes of restriction enzymes.
Figures


References
-
- Bianco PR, Hurley EM (2005) The type I restriction endonuclease EcoR1241, couples ATP hydrolysis to bidirectional DNA translocation. J Mol Biol 352: 837–859 - PubMed
-
- Bist P, Sistla S, Krishnamurthy V, Acharya A, Chandrakala B, Rao DN (2001) S-adenosyl-L-methionine is required for DNA cleavage by type III restriction enzymes. J Mol Biol 310: 93–109 - PubMed
-
- Bourniquel AA, Bickle TA (2002) Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84: 1047–1059 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

