Phenylpyruvate tautomerase activity of trans-3-chloroacrylic acid dehalogenase: evidence for an enol intermediate in the dehalogenase reaction?
- PMID: 17661448
- PMCID: PMC2531067
- DOI: 10.1021/bi7007189
Phenylpyruvate tautomerase activity of trans-3-chloroacrylic acid dehalogenase: evidence for an enol intermediate in the dehalogenase reaction?
Abstract
The enzymatic conversion of cis- or trans-3-chloroacrylic acid to malonate semialdehyde is a key step in the bacterial degradation of the nematocide 1,3-dichloropropene. Two mechanisms have been proposed for the isomer-specific hydrolytic dehalogenases, cis- and trans-3-chloroacrylic acid dehalogenase (cis-CaaD and CaaD, respectively), responsible for this step. In one mechanism, the enol isomer of malonate semialdehyde is produced by the alpha,beta-elimination of HCl from an initial halohydrin species. Phenylenolpyruvate has now been found to be a substrate for CaaD with a kcat/Km value that approaches the one determined for the CaaD reaction using trans-3-chloroacrylate. Moreover, the reaction is stereoselective, generating the 3S isomer of [3-2H]phenylpyruvate in a 1.8:1 ratio in 2H2O. These two observations and a kinetic analysis of active site mutants of CaaD suggest that the active site of CaaD is responsible for the phenylpyruvate tautomerase (PPT) activity. The activity is a striking example of catalytic promiscuity and could reflect the presence of an enol intermediate in CaaD-mediated dehalogenation of trans-3-chloroacrylate. CaaD and cis-CaaD represent different families in the tautomerase superfamily, a group of structurally homologous proteins characterized by a core beta-alpha-beta building block and a catalytic Pro-1. The eukaryotic immunoregulatory protein known as macrophage migration inhibitory factor (MIF), also a tautomerase superfamily member, exhibits a PPT activity, but the biological relevance is unknown. In addition to the mechanistic implications, these results establish a functional link between CaaD and the superfamily tautomerases, highlight the catalytic and binding promiscuity of the beta-alpha-beta scaffold, and suggest that the PPT activity of MIF could reflect a partial reaction in an unknown MIF-catalyzed reaction.
Figures


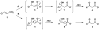


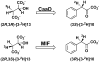

References
-
- Copley SD. Microbial dehalogenases. In: Barton D, Nakanishi K, editors. Comprehensive Natural Products Chemistry. Vol. 5. Elsevier; Amsterdam: 1999. pp. 401–422.
-
- Hartmans S, Jansen MW, van der Werf MJ, de Bont JAM. Bacterial metabolism of 3-chloroacrylic acid. J Gen Microbiol. 1991;137:2025–2032. - PubMed
-
- van Hylckama Vlieg JET, Janssen DB. Bacterial degradation of 3-chloroacrylic acid and the characterization of cis- and trans-specific dehalogenases. Biodegradation. 1992;2:139–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

