A metabolic sensor governing cell size in bacteria
- PMID: 17662947
- PMCID: PMC1971218
- DOI: 10.1016/j.cell.2007.05.043
A metabolic sensor governing cell size in bacteria
Abstract
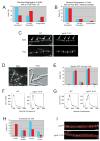
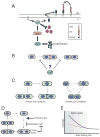
Nutrient availability is one of the strongest determinants of cell size. When grown in rich media, single-celled organisms such as yeast and bacteria can be up to twice the size of their slow-growing counterparts. The ability to modulate size in a nutrient-dependent manner requires cells to: (1) detect when they have reached the appropriate mass for a given growth rate and (2) transmit this information to the division apparatus. We report the identification of a metabolic sensor that couples nutritional availability to division in Bacillus subtilis. A key component of this sensor is an effector, UgtP, which localizes to the division site in a nutrient-dependent manner and inhibits assembly of the tubulin-like cell division protein FtsZ. This sensor serves to maintain a constant ratio of FtsZ rings to cell length regardless of growth rate and ensures that cells reach the appropriate mass and complete chromosome segregation prior to cytokinesis.
Figures
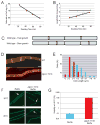
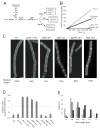
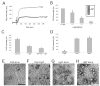
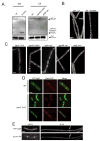
Comment in
-
A sweet sensor for size-conscious bacteria.Cell. 2007 Jul 27;130(2):216-8. doi: 10.1016/j.cell.2007.07.011. Cell. 2007. PMID: 17662935
References
-
- Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

