Digital PCR for the molecular detection of fetal chromosomal aneuploidy
- PMID: 17664418
- PMCID: PMC1934923
- DOI: 10.1073/pnas.0705765104
Digital PCR for the molecular detection of fetal chromosomal aneuploidy
Abstract
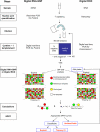
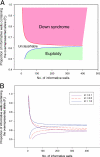
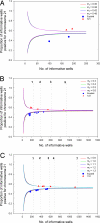
Trisomy 21 is the most common reason that women opt for prenatal diagnosis. Conventional prenatal diagnostic methods involve the sampling of fetal materials by invasive procedures such as amniocentesis. Screening by ultrasonography and biochemical markers have been used to risk-stratify pregnant women before definitive invasive diagnostic procedures. However, these screening methods generally target epiphenomena, such as nuchal translucency, associated with trisomy 21. It would be ideal if noninvasive genetic methods were available for the direct detection of the core pathology of trisomy 21. Here we outline an approach using digital PCR for the noninvasive detection of fetal trisomy 21 by analysis of fetal nucleic acids in maternal plasma. First, we demonstrate the use of digital PCR to determine the allelic imbalance of a SNP on PLAC4 mRNA, a placenta-expressed transcript on chromosome 21, in the maternal plasma of women bearing trisomy 21 fetuses. We named this the digital RNA SNP strategy. Second, we developed a nonpolymorphism-based method for the noninvasive prenatal detection of trisomy 21. We named this the digital relative chromosome dosage (RCD) method. Digital RCD involves the direct assessment of whether the total copy number of chromosome 21 in a sample containing fetal DNA is overrepresented with respect to a reference chromosome. Even without elaborate instrumentation, digital RCD allows the detection of trisomy 21 in samples containing 25% fetal DNA. We applied the sequential probability ratio test to interpret the digital PCR data. Computer simulation and empirical validation confirmed the high accuracy of the disease classification algorithm.
Conflict of interest statement
Conflict of interest statement: Y.M.D.L., F.M.F.L., K.C.A.C., N.B.Y.T., K.C.C., B.C.Y.Z., C.R.C., and R.W.K.C. have filed patent applications on aspects of noninvasive prenatal diagnostics. Y.M.D.L. has equity in Plasmagene Biosciences Limited. C.R.C. has equity in Sequenom, Inc., and is the Chief Scientific Officer of Sequenom, Inc.
Figures
References
-
- Tabor A, Philip J, Madsen M, Bang J, Obel EB, Norgaard-Pedersen B. Lancet. 1986;1:1287–1293. - PubMed
-
- Malone FD, Canick JA, Ball RH, Nyberg DA, Comstock CH, Bukowski R, Berkowitz RL, Gross SJ, Dugoff L, Craigo SD, et al. N Engl J Med. 2005;353:2001–2011. - PubMed
-
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Lancet. 1997;350:485–487. - PubMed
-
- Lo YMD, Chiu RWK. Nat Rev Genet. 2007;8:71–77. (lett) - PubMed
-
- Costa JM, Benachi A, Gautier E. N Engl J Med. 2002;346:1502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

