Gene copy number variation spanning 60 million years of human and primate evolution
- PMID: 17666543
- PMCID: PMC1950895
- DOI: 10.1101/gr.6557307
Gene copy number variation spanning 60 million years of human and primate evolution
Abstract
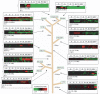
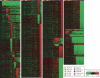
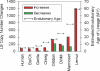
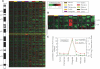
Given the evolutionary importance of gene duplication to the emergence of species-specific traits, we have extended the application of cDNA array-based comparative genomic hybridization (aCGH) to survey gene duplications and losses genome-wide across 10 primate species, including human. Using human cDNA arrays that contained 41,126 cDNAs, corresponding to 24,473 unique human genes, we identified 4159 genes that likely represent most of the major lineage-specific gene copy number gains and losses that have occurred in these species over the past 60 million years. We analyzed 1,233,780 gene-to-gene data points and found that gene gains typically outnumbered losses (ratio of gains/losses = 2.34) and these frequently cluster in complex and dynamic genomic regions that are likely to serve as gene nurseries. Almost one-third of all human genes (6696) exhibit an aCGH- predicted change in copy number in one or more of these species, and within-species gene amplification is also evident. Many of the genes identified here are likely to be important to lineage-specific traits including, for example, human-specific duplications of the AQP7 gene, which represent intriguing candidates to underlie the key physiological adaptations in thermoregulation and energy utilization that permitted human endurance running.
Figures

References
-
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
-
- Bond J., Woods C.G., Woods C.G. Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 2006;18:95–101. - PubMed
-
- Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Morrison E.E., Leal G.F., Silva E.O., Leal G.F., Silva E.O., Silva E.O., et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. - PubMed
-
- Bramble D.M., Lieberman D.E., Lieberman D.E. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
