Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium
- PMID: 17668047
- PMCID: PMC1924603
- DOI: 10.1371/journal.pone.0000659
Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium
Abstract
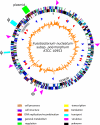
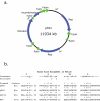
Fusobacterium nucleatum is a prominent member of the oral microbiota and is a common cause of human infection. F. nucleatum includes five subspecies: polymorphum, nucleatum, vincentii, fusiforme, and animalis. F. nucleatum subsp. polymorphum ATCC 10953 has been well characterized phenotypically and, in contrast to previously sequenced strains, is amenable to gene transfer. We sequenced and annotated the 2,429,698 bp genome of F. nucleatum subsp. polymorphum ATCC 10953. Plasmid pFN3 from the strain was also sequenced and analyzed. When compared to the other two available fusobacterial genomes (F. nucleatum subsp. nucleatum, and F. nucleatum subsp. vincentii) 627 open reading frames unique to F. nucleatum subsp. polymorphum ATCC 10953 were identified. A large percentage of these mapped within one of 28 regions or islands containing five or more genes. Seventeen percent of the clustered proteins that demonstrated similarity were most similar to proteins from the clostridia, with others being most similar to proteins from other gram-positive organisms such as Bacillus and Streptococcus. A ten kilobase region homologous to the Salmonella typhimurium propanediol utilization locus was identified, as was a prophage and integrated conjugal plasmid. The genome contains five composite ribozyme/transposons, similar to the CdISt IStrons described in Clostridium difficile. IStrons are not present in the other fusobacterial genomes. These findings indicate that F. nucleatum subsp. polymorphum is proficient at horizontal gene transfer and that exchange with the Firmicutes, particularly the Clostridia, is common.
Conflict of interest statement
Figures



References
-
- Ximénez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27:648–657. - PubMed
-
- Kononen E. Development of oral bacterial flora in young children. Ann Med. 2000;32:107–112. - PubMed
-
- Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiol. 2002;148:467–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

