Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance
- PMID: 17668376
- PMCID: PMC1950802
- DOI: 10.1086/519248
Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance
Abstract
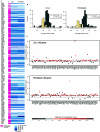
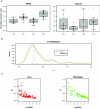
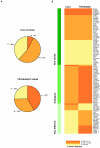
Down syndrome (DS) is characterized by extensive phenotypic variability, with most traits occurring in only a fraction of affected individuals. Substantial gene-expression variation is present among unaffected individuals, and this variation has a strong genetic component. Since DS is caused by genomic-dosage imbalance, we hypothesize that gene-expression variation of human chromosome 21 (HSA21) genes in individuals with DS has an impact on the phenotypic variability among affected individuals. We studied gene-expression variation in 14 lymphoblastoid and 17 fibroblast cell lines from individuals with DS and an equal number of controls. Gene expression was assayed using quantitative real-time polymerase chain reaction on 100 and 106 HSA21 genes and 23 and 26 non-HSA21 genes in lymphoblastoid and fibroblast cell lines, respectively. Surprisingly, only 39% and 62% of HSA21 genes in lymphoblastoid and fibroblast cells, respectively, showed a statistically significant difference between DS and normal samples, although the average up-regulation of HSA21 genes was close to the expected 1.5-fold in both cell types. Gene-expression variation in DS and normal samples was evaluated using the Kolmogorov-Smirnov test. According to the degree of overlap in expression levels, we classified all genes into 3 groups: (A) nonoverlapping, (B) partially overlapping, and (C) extensively overlapping expression distributions between normal and DS samples. We hypothesize that, in each cell type, group A genes are the most dosage sensitive and are most likely involved in the constant DS traits, group B genes might be involved in variable DS traits, and group C genes are not dosage sensitive and are least likely to participate in DS pathological phenotypes. This study provides the first extensive data set on HSA21 gene-expression variation in DS and underscores its role in modulating the outcome of gene-dosage imbalance.
Figures


References
Web Resources
-
- Coriell Institute for Medical Research, http://www.coriell.org/
-
- Ensembl, http://www.ensembl.org/
-
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for trisomy 21)
-
- The R Project for Statistical Computing, http://www.r-project.org
References
-
- Epstein CJ (ed) (1989) Down syndrome, trisomy 21. In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. Metabolic basis of inherited disease. McGraw-Hill, New York, pp 291–326
-
- Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, Eppens B, Saran NG, Pletcher MT, South ST, Stetten G, et al (2004) Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res 14:1258–126710.1101/gr.1951304 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

