Recent advances in the search for antiviral agents against human papillomaviruses
- PMID: 17668552
- PMCID: PMC4646640
Recent advances in the search for antiviral agents against human papillomaviruses
Abstract
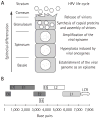
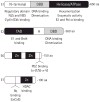
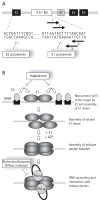
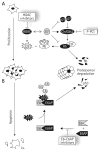
Infection by human papillomavirus (HPV) is extremely common and associated with the development of benign warts or malignant lesions of the skin and mucosa. Infection by a high-risk (oncogenic) anogenital HPV type, most often through sexual contacts, is the starting point of virtually all cases of cervical cancers and the majority of anal cancers. The same viral types are also increasingly being linked with a subset of head-and-neck and non-melanoma skin cancers. Although prophylactic vaccines are now available to protect against the four types most commonly found in cervical and anal cancers (HPV16 and HPV18) and anogenital warts (HPV6 and HPV11), these neither protect against all genital HPVs nor are of therapeutic utility for already infected patients. Thus, the need for antiviral agents to treat HPV-associated diseases remains great, but none currently exist. This article reviews the recent progress made towards the development of antiviral agents to treat HPV infections, from target identification and validation to the discovery of lead compounds with therapeutic potential. Emphasis has been placed on novel low-molecular-weight compounds that antagonize HPV proteins or, alternatively, inhibit cellular proteins which have been usurped by papillomaviruses and are mediating their pathogenic effects.
Figures
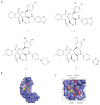
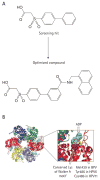
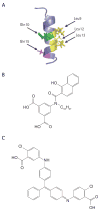
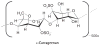
References
-
- Daling JR, Madeleine MM, Johnson LG, et al. Human papillomavirus, smoking, and sexual practices in the etiology of anal cancer. Cancer. 2004;101:270–280. - PubMed
-
- Frisch M, Fenger C, van den Brule AJ, et al. Variants of squamous cell carcinoma of the anal canal and perianal skin and their relation to human papillomaviruses. Cancer Res. 1999;59:753–757. - PubMed
-
- Syrjanen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32(Suppl 1):S59–S66. - PubMed
-
- Akgul B, Cooke JC, Storey A. HPV-associated skin disease. J Pathol. 2006;208:165–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
