High-frequency reversion of geminivirus replication protein mutants during infection
- PMID: 17670823
- PMCID: PMC2045516
- DOI: 10.1128/JVI.00925-07
High-frequency reversion of geminivirus replication protein mutants during infection
Abstract
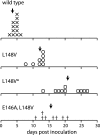
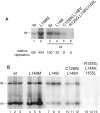
The geminivirus replication protein AL1 interacts with retinoblastoma-related protein (RBR), a key regulator of the plant division cell cycle, to induce conditions permissive for viral DNA replication. Previous studies of tomato golden mosaic virus (TGMV) AL1 showed that amino acid L148 in the conserved helix 4 motif is critical for RBR binding. In this work, we examined the effect of an L148V mutation on TGMV replication in tobacco cells and during infection of Nicotiana benthamiana plants. The L148V mutant replicated 100 times less efficiently than wild-type TGMV in protoplasts but produced severe symptoms that were delayed compared to those of wild-type infection in plants. Analysis of progeny viruses revealed that the L148V mutation reverted at 100% frequency in planta to methionine, leucine, isoleucine, or a second-site mutation depending on the valine codon in the initial DNA sequence. Similar results were seen with another geminivirus, cabbage leaf curl virus (CaLCuV), carrying an L145A mutation in the equivalent residue. Valine was the predominant amino acid recovered from N. benthamiana plants inoculated with the CaLCuV L145A mutant, while threonine was the major residue in Arabidopsis thaliana plants. Together, these data demonstrated that there is strong selection for reversion of the TGMV L148V and CaLCuV L145A mutations but that the nature of the selected revertants is influenced by both the viral background and host components. These data also suggested that high mutation rates contribute to the rapid evolution of geminivirus genomes in plants.
Figures





Similar articles
-
A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein.J Virol. 2004 May;78(9):4817-26. doi: 10.1128/jvi.78.9.4817-4826.2004. J Virol. 2004. PMID: 15078963 Free PMC article.
-
Peptide aptamers that bind to a geminivirus replication protein interfere with viral replication in plant cells.J Virol. 2006 Jun;80(12):5841-53. doi: 10.1128/JVI.02698-05. J Virol. 2006. PMID: 16731923 Free PMC article.
-
Geminivirus replication origins have a modular organization.Plant Cell. 1994 Mar;6(3):405-16. doi: 10.1105/tpc.6.3.405. Plant Cell. 1994. PMID: 8180499 Free PMC article.
-
Geminiviruses and the plant cell cycle.Plant Mol Biol. 2000 Aug;43(5-6):763-72. doi: 10.1023/a:1006462028363. Plant Mol Biol. 2000. PMID: 11089875 Review.
-
Insights into the functional characteristics of geminivirus rolling-circle replication initiator protein and its interaction with host factors affecting viral DNA replication.Arch Virol. 2015 Feb;160(2):375-87. doi: 10.1007/s00705-014-2297-7. Epub 2014 Dec 2. Arch Virol. 2015. PMID: 25449306 Review.
Cited by
-
Adaptive evolution by recombination is not associated with increased mutation rates in Maize streak virus.BMC Evol Biol. 2012 Dec 27;12:252. doi: 10.1186/1471-2148-12-252. BMC Evol Biol. 2012. PMID: 23268599 Free PMC article.
-
Sucrose Nonfermenting 1-Related Protein Kinase 1 Phosphorylates a Geminivirus Rep Protein to Impair Viral Replication and Infection.Plant Physiol. 2018 Sep;178(1):372-389. doi: 10.1104/pp.18.00268. Epub 2018 Jul 13. Plant Physiol. 2018. PMID: 30006378 Free PMC article.
-
Roles of plant retinoblastoma protein: cell cycle and beyond.EMBO J. 2020 Oct 1;39(19):e105802. doi: 10.15252/embj.2020105802. Epub 2020 Aug 31. EMBO J. 2020. PMID: 32865261 Free PMC article. Review.
-
Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense.Elife. 2015 Sep 7;4:e06671. doi: 10.7554/eLife.06671. Elife. 2015. PMID: 26344546 Free PMC article.
-
Identification of the Viral Determinant of Hypovirulence and Host Range in Sclerotiniaceae of a Genomovirus Reconstructed from the Plant Metagenome.J Virol. 2021 Aug 10;95(17):e0026421. doi: 10.1128/JVI.00264-21. Epub 2021 Aug 10. J Virol. 2021. PMID: 34132570 Free PMC article.
References
-
- Amin, I., S. Mansoor, L. Amrao, M. Hussain, S. Irum, Y. Zafar, S. E. Bull, and R. W. Briddon. 2006. Mobilisation into cotton and spread of a recombinant Cotton leaf curl disease satellite. Arch. Virol. 151:2055-2065. - PubMed
-
- Arguello-Astorga, G. R., and R. Ruiz-Medrano. 2001. An iteron-related domain is associated to motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Arch. Virol. 146:1465-1485. - PubMed
-
- Bernard, H. U. 1994. Coevolution of papillomaviruses with human populations. Trends Microbiol. 2:140-143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

