The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA
- PMID: 17670826
- PMCID: PMC2045548
- DOI: 10.1128/JVI.00162-07
The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA
Abstract
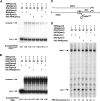
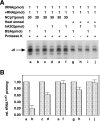
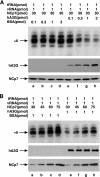
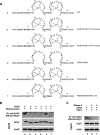
Human immunodeficiency virus type 1 (HIV-1) containing human APOBEC3G (hA3G) has a reduced ability to produce viral DNA in newly infected cells. At least part of this hA3G-facilitated inhibition is due to a cytidine deamination-independent reduction in the ability to initiate reverse transcription. HIV-1 nucleocapsid (NCp7) is required both for the incorporation of hA3G into virions and for the annealing between viral RNA and tRNA(3)(Lys), the primer tRNA for reverse transcription. Herein we present evidence that the interaction of hA3G with nucleocapsid is required for the inhibition of reverse transcription initiation. A tRNA(3)(Lys) priming complex was produced in vitro by the NCp7-facilitated annealing of tRNA(3)(Lys) to synthetic viral RNA in the absence or presence of hA3G. The effect of hA3G on the annealing of tRNA(3)(Lys) to viral RNA and the ability of tRNA(3)(Lys) to initiate reverse transcription was measured. Our results show the following. (i) Electrophoretic band shift and primer binding site assays show that hA3G reduces the annealing of tRNA(3)(Lys) 44 and 60%, respectively, but does not disrupt the annealed complex once formed. (ii) hA3G inhibits tRNA(3)(Lys) priming 70 to 80%. (iii) Inhibition of tRNA(3)(Lys) priming by hA3G requires an interaction between hA3G and NCp7 during annealing. Thus, annealing of tRNA(3)(Lys) is insensitive to hA3G inhibition when facilitated by a zinc finger mutant of NCp7 unable to interact with hA3G. NCp7-independent annealing of DNA to viral RNA also is insensitive to hA3G inhibition. These results indicate that hA3G does not sterically block tRNA(3)(Lys) annealing by binding to viral RNA. Annealing and priming are not affected by another RNA binding protein, QKI-6.
Figures

Similar articles
-
Roles of Gag and NCp7 in facilitating tRNA(Lys)(3) Annealing to viral RNA in human immunodeficiency virus type 1.J Virol. 2009 Aug;83(16):8099-107. doi: 10.1128/JVI.00488-09. Epub 2009 Jun 3. J Virol. 2009. PMID: 19494012 Free PMC article.
-
Human immunodeficiency virus Type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA(Lys3) in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site.J Virol. 1996 Aug;70(8):4996-5004. doi: 10.1128/JVI.70.8.4996-5004.1996. J Virol. 1996. PMID: 8764006 Free PMC article.
-
Inhibition of tRNA₃(Lys)-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication.J Virol. 2006 Dec;80(23):11710-22. doi: 10.1128/JVI.01038-06. Epub 2006 Sep 13. J Virol. 2006. PMID: 16971427 Free PMC article.
-
Aspects of HIV-1 assembly that promote primer tRNA(Lys3) annealing to viral RNA.Virus Res. 2012 Nov;169(2):340-8. doi: 10.1016/j.virusres.2012.06.001. Epub 2012 Jun 12. Virus Res. 2012. PMID: 22698876 Review.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
Cited by
-
Emerging complexities of APOBEC3G action on immunity and viral fitness during HIV infection and treatment.Retrovirology. 2012 Apr 30;9:35. doi: 10.1186/1742-4690-9-35. Retrovirology. 2012. PMID: 22546055 Free PMC article. Review.
-
Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif.J Virol. 2010 Dec;84(24):12599-608. doi: 10.1128/JVI.01437-10. Epub 2010 Oct 13. J Virol. 2010. PMID: 20943965 Free PMC article.
-
Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition.J Virol. 2009 Dec;83(23):12611-21. doi: 10.1128/JVI.01491-09. Epub 2009 Sep 23. J Virol. 2009. PMID: 19776130 Free PMC article.
-
Nucleocapsid mutations turn HIV-1 into a DNA-containing virus.Nucleic Acids Res. 2008 Apr;36(7):2311-9. doi: 10.1093/nar/gkn069. Epub 2008 Feb 22. Nucleic Acids Res. 2008. PMID: 18296486 Free PMC article.
-
Human APOBEC3 Variations and Viral Infection.Viruses. 2021 Jul 14;13(7):1366. doi: 10.3390/v13071366. Viruses. 2021. PMID: 34372572 Free PMC article. Review.
References
-
- Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. - PubMed
-
- Arts, E. J., X. Li, Z. Gu, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672-14680. - PubMed
-
- Beerens, N., and B. Berkhout. 2000. In vitro studies on tRNA annealing and reverse transcription with mutant HIV-1 RNA templates. J. Biol. Chem. 275:15474-15481. - PubMed
-
- Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672-675. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

