Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts
- PMID: 17670833
- PMCID: PMC2045565
- DOI: 10.1128/JVI.00705-07
Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts
Abstract
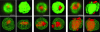
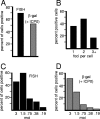
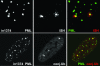
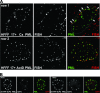
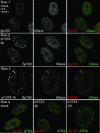
Herpes simplex virus type 1 (HSV-1) genomes become associated with structures related to cellular nuclear substructures known as ND10 or promyelocytic leukemia nuclear bodies during the early stages of lytic infection. This paper describes the relationship between HSV-1 genomes and ND10 in human fibroblasts that maintain the viral genomes in a quiescent state. We report that quiescent HSV-1 genomes detected by fluorescence in situ hybridization (FISH) are associated with enlarged ND10-like structures, frequently such that the FISH-defined viral foci are apparently enveloped within a sphere of PML and other ND10 proteins. The number of FISH viral foci in each quiescently infected cell is concordant with the input multiplicity of infection, with each structure containing no more than a small number of viral genomes. A proportion of the enlarged ND10-like foci in quiescently infected cells contain accumulations of the heterochromatin protein HP1 but not other common markers of heterochromatin such as histone H3 di- or trimethylated on lysine residue 9. Many of the virally induced enlarged ND10-like structures also contain concentrations of conjugated ubiquitin. Quiescent infections can be established in cells that are highly depleted for PML. However, during the initial stages of establishment of a quiescent infection in such cells, other ND10 proteins (Sp100, hDaxx, and ATRX) are recruited into virally induced foci that are likely to be associated with HSV-1 genomes. These observations illustrate that the intimate connections between HSV-1 genomes and ND10 that occur during lytic infection also extend to quiescent infections.
Figures


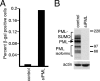
References
-
- Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. - PubMed
-
- Davison, M. J., V. G. Preston, and D. J. McGeoch. 1984. Determination of the sequence alteration in the DNA of the herpes simplex virus type 1 temperature-sensitive mutant ts K. J. Gen. Virol. 65:859-863. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

