p21 transcription is regulated by differential localization of histone H2A.Z
- PMID: 17671089
- PMCID: PMC1935026
- DOI: 10.1101/gad.1545707
p21 transcription is regulated by differential localization of histone H2A.Z
Abstract
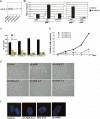
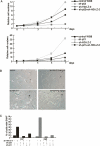
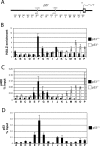
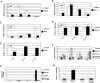
In yeast cells, H2A.Z regulates transcription and is globally associated within a few nucleosomes of the initiator regions of numerous promoters. H2A.Z is deposited at these loci by an ATP-dependent complex, Swr1.com. Here we show that H2A.Z suppresses the p53 --> p21 transcription and senescence responses. Upon DNA damage, H2A.Z is first evicted from the p21 promoter, followed by the recruitment of the Tip60 histone acetyltransferase to activate p21 transcription. p400, a human Swr1 homolog, is required for the localization of H2A.Z, and largely colocalizes with H2A.Z at multiple promoters investigated. Notably, the presence of sequence-specific transcription factors, such as p53 and Myc, provides positioning cues that direct the location of H2A.Z-containing nucleosomes within these promoters. Collectively, this study strongly suggests that certain sequence-specific transcription factors regulate transcription, in part, by preferentially positioning histone variant H2A.Z within chromatin. This H2A.Z-centered process is part of an epigenetic process for modulating gene expression.
Figures


References
-
- Allis C.D., Glover C.V., Bowen J.K., Gorovsky M.A., Glover C.V., Bowen J.K., Gorovsky M.A., Bowen J.K., Gorovsky M.A., Gorovsky M.A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980;20:609–617. - PubMed
-
- Bakshi R., Mehta A.K., Sharma R., Maiti S., Pasha S., Brahmachari V., Mehta A.K., Sharma R., Maiti S., Pasha S., Brahmachari V., Sharma R., Maiti S., Pasha S., Brahmachari V., Maiti S., Pasha S., Brahmachari V., Pasha S., Brahmachari V., Brahmachari V. Characterization of a human SWI2/SNF2 like protein hINO80: Demonstration of catalytic and DNA binding activity. Biochem. Biophys. Res. Commun. 2006;339:313–320. - PubMed
-
- Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Madiredjo M., Nijkamp W., Weigelt B., Nijkamp W., Weigelt B., Weigelt B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. - PubMed
-
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Dorken B., Jenuwein T., Schmitt C.A., Jenuwein T., Schmitt C.A., Schmitt C.A. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
