Heritability of alternative splicing in the human genome
- PMID: 17671095
- PMCID: PMC1933514
- DOI: 10.1101/gr.6281007
Heritability of alternative splicing in the human genome
Abstract
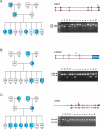
Alternative pre-mRNA splicing increases proteomic diversity and provides a potential mechanism underlying both phenotypic diversity and susceptibility to genetic disorders in human populations. To investigate the variation in splicing among humans on a genome-wide scale, we use a comprehensive exon-targeted microarray to examine alternative splicing in lymphoblastoid cell lines (LCLs) derived from the CEPH HapMap population. We show the identification of transcripts containing sequence verified exon skipping, intron retention, and cryptic splice site usage that are specific between individuals. A number of novel alternative splicing events with no previous annotations in either the RefSeq and EST databases were identified, indicating that we are able to discover de novo splicing events. Using family-based linkage analysis, we demonstrate Mendelian inheritance and segregation of specific splice isoforms with regulatory haplotypes for three genes: OAS1, CAST, and CRTAP. Allelic association was further used to identify individual SNPs or regulatory haplotype blocks linked to the alternative splicing event, taking advantage of the high-resolution genotype information from the CEPH HapMap population. In one candidate, we identified a regulatory polymorphism that disrupts a 5' splice site of an exon in the CAST gene, resulting in its exclusion in the mutant allele. This report illustrates that our approach can detect both annotated and novel alternatively spliced variants, and that such variation among individuals is heritable and genetically controlled.
Figures



References
-
- Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R., Cherny S.S., Cookson W.O., Cardon L.R., Cookson W.O., Cardon L.R., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. - PubMed
-
- Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Daly M.J., Donnelly P., Consortium I.H., Donnelly P., Consortium I.H., Consortium I.H. A haplotype map of the human genome. Nature. 2005;437:1299–1320. - PMC - PubMed
-
- Ben-Ari S., Toiber D., Sas A.S., Soreq H., Ben-Shaul Y., Toiber D., Sas A.S., Soreq H., Ben-Shaul Y., Sas A.S., Soreq H., Ben-Shaul Y., Soreq H., Ben-Shaul Y., Ben-Shaul Y. Modulated splicing-associated gene expression in P19 cells expressing distinct acetylcholinesterase splice variants. J. Neurochem. 2006;97 (Suppl 1):24–34. - PubMed
-
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300.
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
