Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1
- PMID: 17671232
- PMCID: PMC2077318
- DOI: 10.1182/blood-2007-02-073486
Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1
Abstract
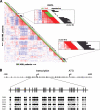
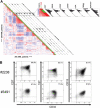
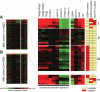
Gene expression profiling of acute myeloid leukemia (AML) allows the discovery of previously unrecognized molecular entities. Here, we identified a specific subgroup of AML, defined by an expression profile resembling that of AMLs with mutations in the myeloid transcription factor CCAAT/enhancer-binding protein alpha (C/EBPalpha), while lacking such mutations. We found that in these leukemias, the CEBPA gene was silenced, which was associated with frequent promoter hypermethylation. The leukemias phenotypically showed aberrant expression of T-cell genes, of which CD7 was most consistent. We identified 2 mechanisms that may contribute to this phenotype. First, absence of Cebpa led to up-regulation of specific T-cell transcripts (ie, Cd7 and Lck) in hematopoietic stem cells isolated from conditional Cebpa knockout mice. Second, the enhanced expression of TRIB2, which we identify here as a direct target of the T-cell commitment factor NOTCH1, suggested aberrantly activated Notch signaling. Putatively activating NOTCH1 mutations were found in several specimens of the newly identified subgroup, while a large set of control AMLs was mutation negative. A gene expression prediction signature allowed the detection of similar cases of leukemia in independent series of AML.
Figures


Comment in
-
C/EBPA methylation is common in T-ALL but not in M0 AML.Blood. 2009 Feb 19;113(8):1864-6; author reply 1866. doi: 10.1182/blood-2008-09-176909. Blood. 2009. PMID: 19228936 No abstract available.
References
-
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. - PubMed
-
- Valk PJ, Delwel R, Lowenberg B. Gene expression profiling in acute myeloid leukemia. Curr Opin Hematol. 2005;12:76–81. - PubMed
-
- Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. - PubMed
-
- Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. - PubMed
-
- Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

