Functional atlas of the integrin adhesome
- PMID: 17671451
- PMCID: PMC2735470
- DOI: 10.1038/ncb0807-858
Functional atlas of the integrin adhesome
Abstract
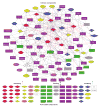
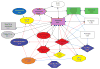
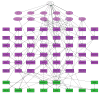
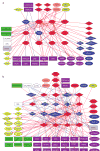
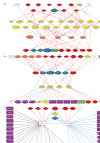
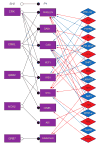
A detailed depiction of the 'integrin adhesome', consisting of a complex network of 156 components linked together and modified by 690 interactions is presented. Different views of the network reveal several functional 'subnets' that are involved in switching on or off many of the molecular interactions within the network, consequently affecting cell adhesion, migration and cytoskeletal organization. Examination of the adhesome network motifs reveals a relatively small number of key motifs, dominated by three-component complexes in which a scaffolding molecule recruits both a signalling molecule and its downstream target. We discuss the role of the different network modules in regulating the structural and signalling functions of cell-matrix adhesions.
Figures
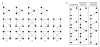
References
-
- Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. - PubMed
-
- Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix — cytoskeleton crosstalk. Nature Rev Mol Cell Biol. 2001;2:793–805. - PubMed
-
- Critchley DR, et al. Integrin-mediated cell adhesion: the cytoskeletal connection. Biochem Soc Symp. 1999;65:79–99. - PubMed
-
- Geiger B, Bershadsky A. Exploring the neighborhood: adhesion-coupled cell mechanosensors. Cell. 2002;110:139–142. - PubMed
-
- Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr Opin Cell Biol. 2006;18:472–481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

