Identification of the PIP2-binding site on Kir6.2 by molecular modelling and functional analysis
- PMID: 17673911
- PMCID: PMC1952224
- DOI: 10.1038/sj.emboj.7601809
Identification of the PIP2-binding site on Kir6.2 by molecular modelling and functional analysis
Abstract
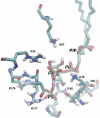
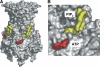
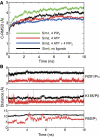
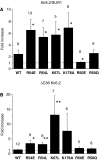
ATP-sensitive potassium (K(ATP)) channels couple cell metabolism to electrical activity by regulating K(+) fluxes across the plasma membrane. Channel closure is facilitated by ATP, which binds to the pore-forming subunit (Kir6.2). Conversely, channel opening is potentiated by phosphoinositol bisphosphate (PIP(2)), which binds to Kir6.2 and reduces channel inhibition by ATP. Here, we use homology modelling and ligand docking to identify the PIP(2)-binding site on Kir6.2. The model is consistent with a large amount of functional data and was further tested by mutagenesis. The fatty acyl tails of PIP(2) lie within the membrane and the head group extends downwards to interact with residues in the N terminus (K39, N41, R54), transmembrane domains (K67) and C terminus (R176, R177, E179, R301) of Kir6.2. Our model suggests how PIP(2) increases channel opening and decreases ATP binding and channel inhibition. It is likely to be applicable to the PIP(2)-binding site of other Kir channels, as the residues identified are conserved and influence PIP(2) sensitivity in other Kir channel family members.
Figures



References
-
- Ashcroft FM (2006) Ion channels and disease: from molecule to malady. Nature 440: 440–447 - PubMed
-
- Baukrowitz T, Schulte U, Oliver D, Herlitze S, Krauter T, Tucker SJ, Ruppersberg JP, Fakler B (1998) PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science 282: 1141–1144 - PubMed
-
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81: 3684–3690
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

