Genes under positive selection in Escherichia coli
- PMID: 17675366
- PMCID: PMC1950902
- DOI: 10.1101/gr.6254707
Genes under positive selection in Escherichia coli
Abstract
We used a comparative genomics approach to identify genes that are under positive selection in six strains of Escherichia coli and Shigella flexneri, including five strains that are human pathogens. We find that positive selection targets a wide range of different functions in the E. coli genome, including cell surface proteins such as beta barrel porins, presumably because of the involvement of these genes in evolutionary arms races with other bacteria, phages, and/or the host immune system. Structural mapping of positively selected sites on trans-membrane beta barrel porins reveals that the residues under positive selection occur almost exclusively in the extracellular region of the proteins that are enriched with sites known to be targets of phages, colicins, or the host immune system. More surprisingly, we also find a number of other categories of genes that show very strong evidence for positive selection, such as the enigmatic rhs elements and transposases. Based on structural evidence, we hypothesize that the selection acting on transposases is related to the genomic conflict between transposable elements and the host genome.
Figures
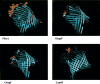

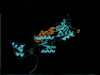
References
-
- Ajioka J., Hartl D.L., Hartl D.L. Population dynamics of transposable elements. In: Berg D., How M.M., How M.M., editors. Mobile DNA. American Society for Microbiology; Washington, DC: 1989. pp. 939–958.
-
- Andersen C., Bachmeyer C., Tauber H., Benz R., Wang J., Michel V., Newton S.M., Hofnung M., Charbit A., Bachmeyer C., Tauber H., Benz R., Wang J., Michel V., Newton S.M., Hofnung M., Charbit A., Tauber H., Benz R., Wang J., Michel V., Newton S.M., Hofnung M., Charbit A., Benz R., Wang J., Michel V., Newton S.M., Hofnung M., Charbit A., Wang J., Michel V., Newton S.M., Hofnung M., Charbit A., Michel V., Newton S.M., Hofnung M., Charbit A., Newton S.M., Hofnung M., Charbit A., Hofnung M., Charbit A., Charbit A. In vivo and in vitro studies of major surface loop deletion mutants of the Escherichia coli K-12 maltoporin: Contribution to maltose and maltooligosaccharide transport and binding. Mol. Microbiol. 1999;32:851–867. - PubMed
-
- Bos C., Lorenzen D., Braun V., Lorenzen D., Braun V., Braun V. Specific in vivo labeling of cell surface-exposed protein loops: Reactive cysteines in the predicted gating loop mark a ferrichrome binding site and a ligand-induced conformational change of the Escherichia coli FhuA protein. J. Bacteriol. 1998;180:605–613. - PMC - PubMed
-
- Braun V., Patzer S.I., Hantke K., Patzer S.I., Hantke K., Hantke K. Ton-dependent colicins and microcins: Modular design and evolution. Biochimie. 2002;84:365–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
