Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome
- PMID: 17675389
- PMCID: PMC2168654
- DOI: 10.1128/JB.00690-07
Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome
Abstract
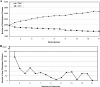
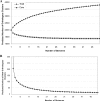
The distributed-genome hypothesis (DGH) states that pathogenic bacteria possess a supragenome that is much larger than the genome of any single bacterium and that these pathogens utilize genetic recombination and a large, noncore set of genes as a means of diversity generation. We sequenced the genomes of eight nasopharyngeal strains of Streptococcus pneumoniae isolated from pediatric patients with upper respiratory symptoms and performed quantitative genomic analyses among these and nine publicly available pneumococcal strains. Coding sequences from all strains were grouped into 3,170 orthologous gene clusters, of which 1,454 (46%) were conserved among all 17 strains. The majority of the gene clusters, 1,716 (54%), were not found in all strains. Genic differences per strain pair ranged from 35 to 629 orthologous clusters, with each strain's genome containing between 21 and 32% noncore genes. The distribution of the orthologous clusters per genome for the 17 strains was entered into the finite-supragenome model, which predicted that (i) the S. pneumoniae supragenome contains more than 5,000 orthologous clusters and (ii) 99% of the orthologous clusters ( approximately 3,000) that are represented in the S. pneumoniae population at frequencies of >or=0.1 can be identified if 33 representative genomes are sequenced. These extensive genic diversity data support the DGH and provide a basis for understanding the great differences in clinical phenotype associated with various pneumococcal strains. When these findings are taken together with previous studies that demonstrated the presence of a supragenome for Streptococcus agalactiae and Haemophilus influenzae, it appears that the possession of a distributed genome is a common host interaction strategy.
Figures



References
-
- Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. - PubMed
-
- Claverys, J. P., and L. S. Havarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-d1814. - PubMed
-
- Daraselia, N., D. Dernovoy, Y. Tian, M. Borodovsky, R. Tatusov, and T. Tatusova. 2003. Reannotation of Shewanella oneidensis genome. OMICS 7:171-175. - PubMed
-
- Del Beccaro, M. A., P. M. Mendelman, A. F. Inglis, M. A. Richardson, N. O. Duncan, C. R. Clausen, and T. L. Stull. 1992. Bacteriology of acute otitis media: a new perspective. J. Pediatr. 120:81-84. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

