A downstream mediator in the growth repression limb of the jasmonate pathway
- PMID: 17675405
- PMCID: PMC2002611
- DOI: 10.1105/tpc.107.050708
A downstream mediator in the growth repression limb of the jasmonate pathway
Abstract
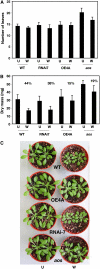
Wounding plant tissues initiates large-scale changes in transcription coupled to growth arrest, allowing resource diversion for defense. These processes are mediated in large part by the potent lipid regulator jasmonic acid (JA). Genes selected from a list of wound-inducible transcripts regulated by the jasmonate pathway were overexpressed in Arabidopsis thaliana, and the transgenic plants were then assayed for sensitivity to methyl jasmonate (MeJA). When grown in the presence of MeJA, the roots of plants overexpressing a gene of unknown function were longer than those of wild-type plants. When transcript levels for this gene, which we named JASMONATE-ASSOCIATED1 (JAS1), were reduced by RNA interference, the plants showed increased sensitivity to MeJA and growth was inhibited. These gain- and loss-of-function assays suggest that this gene acts as a repressor of JA-inhibited growth. An alternative transcript from the gene encoding a second protein isoform with a longer C terminus failed to repress jasmonate sensitivity. This identified a conserved C-terminal sequence in JAS1 and related genes, all of which also contain Zim motifs and many of which are jasmonate-regulated. Both forms of JAS1 were found to localize to the nucleus in transient expression assays. Physiological tests of growth responses after wounding were consistent with the fact that JAS1 is a repressor of JA-regulated growth retardation.
Figures





References
-
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics Int. 11 36–42.
-
- Achard, P., Cheng, H., De Grawe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. - PubMed
-
- Bonaventure, G., Gfeller, A., Proebsting, W.M., Hoerstensteiner, S., Chételat, A., Martinoia, E., and Farmer, E.E. (2007). A gain of function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J. 49 889–898. - PubMed
-
- Browse, J. (2005). Jasmonate: An oxylipin signal with many roles in plants. Vitam. Horm. 72 431–456. - PubMed
-
- Caldelari, D., and Farmer, E.E. (1998). Rapid assays for the coupled cell free generation of oxylipins. Phytochemistry 47 599–604.
NOTE ADDED IN PROOF
-
- Thines et al. (2007) and Chini et al. (2007) have reported finding JAZ proteins that repress jasmonate responses and are degraded in a COI1-dependent manner in jasmonate signaling. Their list includes JAS1, which, in their nomenclature, is JAZ10.
-
- Chini, A., Fonseca, S., Fernández, G., Adie, G., Chico, J.M., Lorenzo, O., García-Casado, G., López-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (July 18, 2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature http://dx.doi.org/10.1038/nature06006. - DOI - PubMed
-
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (July 18, 2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature http://dx.doi.org/10.1038/nature05960. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

