Agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern
- PMID: 17675424
- PMCID: PMC2075002
- DOI: 10.1128/AEM.00608-07
Agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern
Abstract
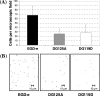
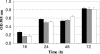
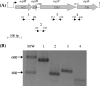
In this study, we investigated the agrBDCA operon in the pathogenic bacterium Listeria monocytogenes EGD-e. In-frame deletion of agrA and agrD resulted in an altered adherence and biofilm formation on abiotic surfaces, suggesting the involvement of the agr system of L. monocytogenes during the early stages of biofilm formation. Real-time PCR experiments indicated that the transcript levels of agrBDCA depended on the stage of biofilm development, since the levels were lower after the initial attachment period than during biofilm growth, whereas transcription during planktonic growth was not growth phase dependent. The mRNA quantification data also suggested that the agr system was autoregulated and pointed to a differential expression of the agr genes during sessile and planktonic growth. Although the reverse transcription-PCR experiments revealed that the four genes were transcribed as a single messenger, chemical half-life and 5' RACE (rapid amplification of cDNA ends) experiments indicated that the full size transcript underwent cleavage followed by degradation of the agrC and agrA transcripts, which suggests a complex regulation of agr transcription.
Figures

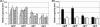


References
-
- Aguado, V., A. I. Vitas, and I. Garcia-Jalon. 2004. Characterization of Listeria monocytogenes and Listeria innocua from a vegetable processing plant by RAPD and REA. Int. J. Food Microbiol. 90:341-347. - PubMed
-
- Allenby, N. E. E., N. O'Connor, Z. Pragai, N. M. Carter, M. Miethke, S. Engelmann, M. Hecker, A. Wipat, A. C. Ward, and C. R. Harwood. 2004. Post-transcriptional regulation of the Bacillus subtilis pst operon encoding a phosphate-specific ABC transporter. Microbiology 150:2619-2628. - PubMed
-
- Allison, D. G., B. Ruiz, C. San Jose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. - PubMed
-
- Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

