SigmaB-dependent and sigmaB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth
- PMID: 17675428
- PMCID: PMC2074989
- DOI: 10.1128/AEM.00714-07
SigmaB-dependent and sigmaB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth
Abstract
The role of the stress response regulator sigma(B) (encoded by sigB) in directing the expression of selected putative and confirmed cold response genes was evaluated using Listeria monocytogenes 10403S and an isogenic DeltasigB mutant, which were either cold shocked at 4 degrees C in brain heart infusion (BHI) broth for up to 30 min or grown at 4 degrees C in BHI for 12 days. Transcript levels of the housekeeping genes rpoB and gap, the sigma(B)-dependent genes opuCA and bsh, and the cold stress genes ltrC, oppA, and fri were measured using quantitative reverse transcriptase PCR. Transcriptional start sites for ltrC, oppA, and fri were determined using rapid amplification of cDNA ends PCR. Centrifugation was found to rapidly induce sigma(B)-dependent transcription, which necessitated the use of centrifugation-independent protocols to evaluate the contributions of sigma(B) to transcription during cold shock. Our data confirmed that transcription of the cold stress genes ltrC and fri is at least partially sigma(B) dependent and experimentally identified a sigma(B)-dependent ltrC promoter. In addition, our data indicate that (i) while sigma(B) activity is induced during 30 min of cold shock, this cold shock does not induce the transcription of sigma(B)-dependent or -independent cold shock genes; (ii) sigma(B) is not required for L. monocytogenes growth at 4 degrees C in BHI; and (iii) transcription of the putative cold stress genes opuCA, fri, and oppA is sigma(B) independent during growth at 4 degrees C, while both bsh and ltrC show growth phase and sigma(B)-dependent transcription during growth at 4 degrees C. We conclude that sigma(B)-dependent and sigma(B)-independent mechanisms contribute to the ability of L. monocytogenes to survive and grow at low temperatures.
Figures
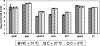

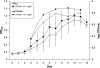

 ) grown at 37°C and for the parent strain and ΔsigB mutant grown at 4°C (▴).
) grown at 37°C and for the parent strain and ΔsigB mutant grown at 4°C (▴).Similar articles
-
Sigma(B)-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes.Microbiology (Reading). 2003 Nov;149(Pt 11):3247-3256. doi: 10.1099/mic.0.26526-0. Microbiology (Reading). 2003. PMID: 14600237
-
SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes.Appl Environ Microbiol. 2006 Aug;72(8):5197-203. doi: 10.1128/AEM.03058-05. Appl Environ Microbiol. 2006. PMID: 16885265 Free PMC article.
-
Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth.J Food Prot. 2008 Feb;71(2):420-5. doi: 10.4315/0362-028x-71.2.420. J Food Prot. 2008. PMID: 18326199 Free PMC article.
-
[Contribution of sigma B to environmental stress tolerance in Listeria monocytogenes--a review].Wei Sheng Wu Xue Bao. 2009 Oct;49(10):1282-8. Wei Sheng Wu Xue Bao. 2009. PMID: 20069872 Review. Chinese.
-
Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism.Future Microbiol. 2019 Jun;14:801-828. doi: 10.2217/fmb-2019-0072. Epub 2019 Jul 4. Future Microbiol. 2019. PMID: 31271064
Cited by
-
Identification of Bacillus cereus genes specifically expressed during growth at low temperatures.Appl Environ Microbiol. 2010 Apr;76(8):2562-73. doi: 10.1128/AEM.02348-09. Epub 2010 Feb 26. Appl Environ Microbiol. 2010. PMID: 20190083 Free PMC article.
-
Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance.Appl Environ Microbiol. 2008 Nov;74(22):6848-58. doi: 10.1128/AEM.00442-08. Epub 2008 Sep 19. Appl Environ Microbiol. 2008. PMID: 18806006 Free PMC article.
-
Transcriptomic and Phenotypic Analyses of the Sigma B-Dependent Characteristics and the Synergism between Sigma B and Sigma L in Listeria monocytogenes EGD-e.Microorganisms. 2020 Oct 23;8(11):1644. doi: 10.3390/microorganisms8111644. Microorganisms. 2020. PMID: 33114171 Free PMC article.
-
The Role of Stress and Stress Adaptations in Determining the Fate of the Bacterial Pathogen Listeria monocytogenes in the Food Chain.Front Microbiol. 2016 Nov 23;7:1865. doi: 10.3389/fmicb.2016.01865. eCollection 2016. Front Microbiol. 2016. PMID: 27933042 Free PMC article. Review.
-
Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology.Virulence. 2021 Dec;12(1):2509-2545. doi: 10.1080/21505594.2021.1975526. Virulence. 2021. PMID: 34612177 Free PMC article.
References
-
- Bayles, D. O., and B. J. Wilkinson. 2000. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett. Appl. Microbiol. 30:23-27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

