Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1
- PMID: 17675441
- PMCID: PMC2074908
- DOI: 10.1128/AEM.00146-07
Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1
Abstract
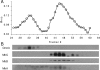
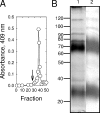
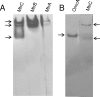
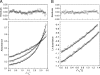
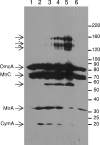
The interaction of proteins implicated in dissimilatory metal reduction by Shewanella oneidensis MR-1 (outer membrane [OM] proteins OmcA, MtrB, and MtrC; OM-associated protein MtrA; periplasmic protein CctA; and cytoplasmic membrane protein CymA) were characterized by protein purification, analytical ultracentrifugation, and cross-linking methods. Five of these proteins are heme proteins, OmcA (83 kDa), MtrC (75 kDa), MtrA (32 kDa), CctA (19 kDa), and CymA (21 kDa), and can be visualized after sodium dodecyl sulfate-polyacrylamide gel electrophoresis by heme staining. We show for the first time that MtrC, MtrA, and MtrB form a 198-kDa complex with a 1:1:1 stoichiometry. These proteins copurify through anion-exchange chromatography, and the purified complex has the ability to reduce multiple forms of Fe(III) and Mn(IV). Additionally, MtrA fractionates with the OM through sucrose density gradient ultracentrifugation, and MtrA comigrates with MtrB in native gels. Protein cross-linking of whole cells with 1% formaldehyde show new heme bands of 160, 151, 136, and 59 kDa. Using antibodies to detect each protein separately, heme proteins OmcA and MtrC were shown to cross-link, yielding the 160-kDa band. Consistent with copurification results, MtrB cross-links with MtrA, forming high-molecular-mass bands of approximately 151 and 136 kDa.
Figures





References
-
- Arslan, E., H. Schulz, R. Zufferey, P. Kunzler, and L. Thony-Meyer. 1998. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251:744-747. - PubMed
-
- Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

