Cloning and transfer of the Salmonella pathogenicity island 2 type III secretion system for studies of a range of gram-negative genera
- PMID: 17675443
- PMCID: PMC2074921
- DOI: 10.1128/AEM.00952-07
Cloning and transfer of the Salmonella pathogenicity island 2 type III secretion system for studies of a range of gram-negative genera
Abstract
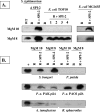
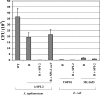
The engineering of bacterial strains with specific phenotypes frequently requires the use of blocks or "cassettes" of genes that act together to perform a desired function. The potential benefits of utilizing type III secretion systems in this regard are becoming increasingly realized since these systems can be used to direct interactions with host cells for beneficial purposes such as vaccine development, anticancer therapies, and targeted protein delivery. However, convenient methods to clone and transfer type III secretion systems for studies of a range of different types of bacteria are lacking. In addition to functional applications, such methods would also reveal important information about the evolution of a given type III secretion system, such as its ability to be expressed and functional outside of the strain of origin. We describe here the cloning of the Salmonella enterica serovar Typhimurium pathogenicity island 2 (SPI-2) type III secretion system onto a vector that can be easily transferred to a range of gram-negative bacterial genera. We found that expression of the cloned SPI-2 system in different Gammaproteobacteria and Alphaproteobacteria (as monitored by SseB protein levels) is dependent on the bacterial strain and growth medium. We also demonstrate that the cloned system is functional for secretion, can direct interactions with macrophages, and can be used as a novel tool to analyze the predicted interaction of SseB with host cells. This work provides a foundation for future applications where the cloned SPI-2 region (or other cloned type III systems) can provide a desired function to an engineered gram-negative strain.
Figures

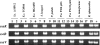



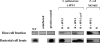
Similar articles
-
Cloning of a functional Salmonella SPI-1 type III secretion system and development of a method to create mutations and epitope fusions in the cloned genes.J Biotechnol. 2006 Mar 23;122(2):147-60. doi: 10.1016/j.jbiotec.2005.09.005. Epub 2005 Oct 25. J Biotechnol. 2006. PMID: 16253373
-
Transfer of the cloned Salmonella SPI-1 type III secretion system and characterization of its expression mechanisms in Gram negative bacteria in comparison with cloned SPI-2.Microbiol Res. 2015 Nov;180:57-64. doi: 10.1016/j.micres.2015.07.006. Epub 2015 Jul 29. Microbiol Res. 2015. PMID: 26505312
-
SsaM and SpiC interact and regulate secretion of Salmonella pathogenicity island 2 type III secretion system effectors and translocators.Mol Microbiol. 2004 Nov;54(3):604-19. doi: 10.1111/j.1365-2958.2004.04297.x. Mol Microbiol. 2004. PMID: 15491354
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
-
Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium.Infect Genet Evol. 2005 Jan;5(1):1-9. doi: 10.1016/j.meegid.2004.07.004. Infect Genet Evol. 2005. PMID: 15567133 Review.
Cited by
-
Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation.PLoS One. 2012;7(3):e33080. doi: 10.1371/journal.pone.0033080. Epub 2012 Mar 8. PLoS One. 2012. PMID: 22412985 Free PMC article.
-
Polyphyletic Nature of Salmonella enterica Serotype Derby and Lineage-Specific Host-Association Revealed by Genome-Wide Analysis.Front Microbiol. 2018 May 17;9:891. doi: 10.3389/fmicb.2018.00891. eCollection 2018. Front Microbiol. 2018. PMID: 29867804 Free PMC article.
-
Bacterial genus-specific tolerance for YdcI expression.Curr Microbiol. 2014 Nov;69(5):640-8. doi: 10.1007/s00284-014-0631-7. Epub 2014 Jun 25. Curr Microbiol. 2014. PMID: 24962596
-
Characterization of the Salmonella enterica serovar Typhimurium ydcI gene, which encodes a conserved DNA binding protein required for full acid stress resistance.J Bacteriol. 2011 May;193(9):2208-17. doi: 10.1128/JB.01335-10. Epub 2011 Mar 11. J Bacteriol. 2011. PMID: 21398541 Free PMC article.
-
Transfer and analysis of Salmonella pdu genes in a range of Gram-negative bacteria demonstrate exogenous microcompartment expression across a variety of species.Microb Biotechnol. 2018 Jan;11(1):199-210. doi: 10.1111/1751-7915.12863. Epub 2017 Oct 2. Microb Biotechnol. 2018. PMID: 28967207 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
-
- Baker, D., G. Church, J. Collins, D. Endy, J. Jacobson, J. Keasling, P. Modrich, C. Smolke, and R. Weiss. 2006. Engineering life: building a fab for biology. Sci. Am. 294:44-51. - PubMed
-
- Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. - PubMed
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

