Characterising alternate splicing and tissue specific expression in the chicken from ESTs
- PMID: 17675868
- PMCID: PMC2266501
- DOI: 10.1159/000103188
Characterising alternate splicing and tissue specific expression in the chicken from ESTs
Abstract
Alternate splicing is believed to produce the greatest diversity in transcriptional complexity and function in eukaryotic species. In this study, we present an analysis of alternative splicing events that occur in the chicken, using the recently sequenced genomic sequence and over 580,000 EST sequences mapped back to the genome. A carefully controlled EST-to-genome mapping pipeline is presented, based around the EXONERATE program using the est2genome model, which also considers several quality control steps to filter out erroneous matches. The data is then used to estimate the level of alternate splicing events with respect to Ensembl predicted transcripts. The EST-genome mappings are characterised at the exon level, in order to classify individual splicing events and provide estimates of novel transcripts not currently annotated by the Ensembl genome database. This is the first large scale analysis of this kind in an avian species, and suggests that chicken displays a similar level of alternate splicing as that found in other higher vertebrates such as human and mouse, both in terms of the number of genes that undergo alternate splicing events, and the average number of transcripts produced per gene. The EST data suggests alternate splicing may occur in some 50-60% of the chicken gene set and with an average of around 2.3 transcripts per gene which undergo this process. The EST data is also used to look at gene and transcript usage in the tissues sequenced in embryonic and adult libraries. Genes which display notable biases were analysed in more detail, including twinfilin-2 and embryonic heavy chain myosin. This also highlights several as yet functionally un-annotated genes which appear to be important in embryonic tissues and also undergo alternate splicing events. The analysis also demonstrates some of the difficulties involved in using EST-based data to annotate transcriptional activity in eukaryotic genes, where a broad spectrum of tissues and a large number of sequenced transcripts are required in order to fully characterise alternate splicing and differential expression.
Copyright 2007 S. Karger AG, Basel.
Figures
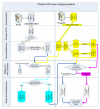
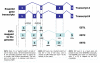

References
-
- Bertaux O, Toselli-Mollereau E, Auffray C, Devignes MD. Alternative usage of 5′ exons in the chicken nerve growth factor gene: refined characterization of a weakly expressed gene. Gene. 2004;334:83–97. - PubMed
-
- Boardman P, Sanz-Ezquerro J, Overton I, Burt D, Bosch E, Fong W, et al. A comprehensive collection of chicken cDNAs. Current Biology. 2002;12:1965–1969. - PubMed
-
- Brown W, Hubbard SJ, Tickle C, Wilson SA. The chicken as a model for large-scale analysis of vertebrate gene function. Nature Rev Genetics. 2003;4:87–98. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

