Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation
- PMID: 17676974
- PMCID: PMC1925137
- DOI: 10.1371/journal.pbio.0050201
Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation
Abstract
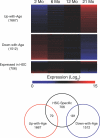
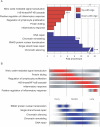
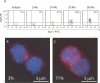
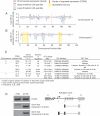
Age-related defects in stem cells can limit proper tissue maintenance and hence contribute to a shortened lifespan. Using highly purified hematopoietic stem cells from mice aged 2 to 21 mo, we demonstrate a deficit in function yet an increase in stem cell number with advancing age. Expression analysis of more than 14,000 genes identified 1,500 that were age-induced and 1,600 that were age-repressed. Genes associated with the stress response, inflammation, and protein aggregation dominated the up-regulated expression profile, while the down-regulated profile was marked by genes involved in the preservation of genomic integrity and chromatin remodeling. Many chromosomal regions showed coordinate loss of transcriptional regulation; an overall increase in transcriptional activity with age and inappropriate expression of genes normally regulated by epigenetic mechanisms was also observed. Hematopoietic stem cells from early-aging mice expressing a mutant p53 allele reveal that aging of stem cells can be uncoupled from aging at an organismal level. These studies show that hematopoietic stem cells are not protected from aging. Instead, loss of epigenetic regulation at the chromatin level may drive both functional attenuation of cells, as well as other manifestations of aging, including the increased propensity for neoplastic transformation.
Conflict of interest statement
Figures


Comment in
-
Mechanisms of aging in bone marrow stem cells.PLoS Biol. 2007 Aug;5(8):e215. doi: 10.1371/journal.pbio.0050215. Epub 2007 Jul 24. PLoS Biol. 2007. PMID: 20076681 Free PMC article. No abstract available.
References
-
- Van Zant G, Liang Y. The role of stem cells in aging. Exp Hematol. 2003;31:659–672. - PubMed
-
- Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nat Med. 1996;2:1011–1016. - PubMed
-
- de Haan G, Van Zant G. Dynamic changes in mouse hematopoietic stem cell numbers during aging. Blood. 1999;93:3294–3301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

