Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells
- PMID: 17676993
- PMCID: PMC1933458
- DOI: 10.1371/journal.pbio.0050218
Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells
Abstract
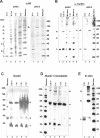
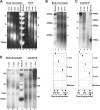
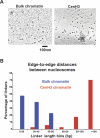
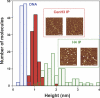
Centromeres, the specialized chromatin structures that are responsible for equal segregation of chromosomes at mitosis, are epigenetically maintained by a centromere-specific histone H3 variant (CenH3). However, the mechanistic basis for centromere maintenance is unknown. We investigated biochemical properties of CenH3 nucleosomes from Drosophila melanogaster cells. Cross-linking of CenH3 nucleosomes identifies heterotypic tetramers containing one copy of CenH3, H2A, H2B, and H4 each. Interphase CenH3 particles display a stable association of approximately 120 DNA base pairs. Purified centromeric nucleosomal arrays have typical "beads-on-a-string" appearance by electron microscopy but appear to resist condensation under physiological conditions. Atomic force microscopy reveals that native CenH3-containing nucleosomes are only half as high as canonical octameric nucleosomes are, confirming that the tetrameric structure detected by cross-linking comprises the entire interphase nucleosome particle. This demonstration of stable half-nucleosomes in vivo provides a possible basis for the instability of centromeric nucleosomes that are deposited in euchromatic regions, which might help maintain centromere identity.
Conflict of interest statement
Figures


References
-
- Amor DJ, Kalitsis P, Sumer H, Choo KH. Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 2004;14:359–368. - PubMed
-
- Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. - PubMed
-
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, et al. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

