Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain
- PMID: 17678855
- PMCID: PMC2039927
- DOI: 10.1016/j.neuron.2007.06.036
Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain
Abstract
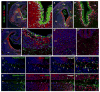
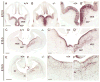
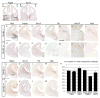
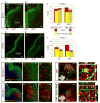
Progenitors within the ventral telencephalon can generate GABAergic neurons and oligodendrocytes, but regulation of the neuron-glial switch is poorly understood. We investigated the combinatorial expression and function of Dlx1&2, Olig2, and Mash1 transcription factors in the ventral telencephalon. We show that Dlx homeobox transcription factors, required for GABAergic interneuron production, repress oligodendrocyte precursor cell (OPC) formation by acting on a common progenitor to determine neuronal versus oligodendroglial cell fate acquisition. We demonstrate that Dlx1&2 negatively regulate Olig2-dependant OPC formation and that Mash1 promotes OPC formation by restricting the number of Dlx+ progenitors. Progenitors transplanted from Dlx1&2 mutant ventral telencephalon into newborn wild-type mice do not produce neurons but differentiate into myelinating oligodendrocytes that survive into adulthood. Our results identify another role for Dlx genes as modulators of neuron versus oligodendrocyte development in the ventral embryonic forebrain.
Figures



References
-
- Agius E, Soukkarieh C, Danesin C, Kan P, Takebayashi H, Soula C, Cochard P. Converse control of oligodendrocyte and astrocyte lineage development by Sonic hedgehog in the chick spinal cord. Dev Biol. 2004;270:308–321. - PubMed
-
- Alberta JA, Park SK, Mora J, Yuk D, Pawlitzky I, Iannarelli P, Vartanian T, Stiles CD, Rowitch DH. Sonic hedgehog is required during an early phase of oligodendrocyte development in mammalian brain. Mol Cell Neurosci. 2001;18:434–441. - PubMed
-
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9:646–654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

