An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae
- PMID: 17679088
- PMCID: PMC2144737
- DOI: 10.1016/j.molcel.2007.06.012
An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae
Abstract
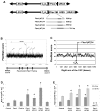
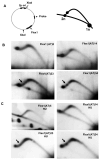
Common fragile sites are regions of human chromosomes prone to breakage. Fragile site FRA16D spans the WWOX/FOR tumor suppressor gene and has been linked to cancer-causing deletions and translocations. Using a genetic assay in yeast, we found that a short AT-rich region (Flex1) within FRA16D increases chromosome fragility, whereas three other sequences within FRA16D do not. To our knowledge, this is the first identification of a sequence element within a common fragile site that increases chromosome fragility. The fragility of Flex1 was exacerbated by the absence of Rad52 or the presence of hydroxyurea. Flex1 contains a polymorphic AT repeat predicted to form a DNA structure, and two-dimensional gel analysis showed accumulation of stalled replication forks at the Flex1 sequence that was dependent on AT length. Our data suggest that the FRA16D Flex1 sequence causes increased chromosome breakage by forming secondary structures that stall replication fork progression.
Figures





References
-
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. - PubMed
-
- Bowater R, Aboul-ela F, Lilley DM. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991;30:11495–11506. - PubMed
-
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR Regulates Fragile Site Stability. Cell. 2002;111:779–789. - PubMed
-
- Cha RS, Kleckner N. ATR Homolog Mec1 Promotes Fork Progression, Thus Averting Breaks in Replication Slow Zones. Science. 2002;297:602–606. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

