Structural proteomics of the SARS coronavirus: a model response to emerging infectious diseases
- PMID: 17680348
- PMCID: PMC7088133
- DOI: 10.1007/s10969-007-9024-5
Structural proteomics of the SARS coronavirus: a model response to emerging infectious diseases
Abstract
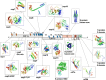
A number of structural genomics/proteomics initiatives are focused on bacterial or viral pathogens. In this article, we will review the progress of structural proteomics initiatives targeting the SARS coronavirus (SARS-CoV), the etiological agent of the 2003 worldwide epidemic that culminated in approximately 8,000 cases and 800 deaths. The SARS-CoV genome encodes 28 proteins in three distinct classes, many of them with unknown function and sharing low similarity to other proteins. The structures of 16 SARS-CoV proteins or functional domains have been determined to date. Remarkably, eight of these 16 proteins or functional domains have novel folds, indicating the uniqueness of the coronavirus proteins. The results of SARS-CoV structural proteomics initiatives will have several profound biological impacts, including elucidation of the structure-function relationships of coronavirus proteins; identification of targets for the design of anti-viral compounds against SARS-CoV and other coronaviruses; and addition of new protein folds to the fold space, with further understanding of the structure-function relationships for several new protein families. We discuss the use of structural proteomics in response to emerging infectious diseases such as SARS-CoV and to increase preparedness against future emerging coronaviruses.
Figures
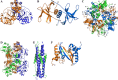

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

