Forkhead transcription factors regulate mosquito reproduction
- PMID: 17681238
- PMCID: PMC2441594
- DOI: 10.1016/j.ibmb.2007.05.008
Forkhead transcription factors regulate mosquito reproduction
Abstract
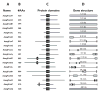
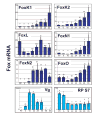
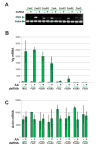
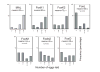
Forkhead-box (Fox) genes encode a family of transcription factors defined by a 'winged helix' DNA-binding domain. In this study we aimed to identify Fox factors that are expressed within the fat body of the yellow fever mosquito Aedes aegypti, and determine whether any of these are involved in the regulation of mosquito yolk protein gene expression. The Ae. aegypti genome contains 18 loci that encode putative Fox factors. Our stringent cladistic analysis has profound implications for the use of Fox genes as phylogenetic markers. Twelve Ae. aegypti Fox genes are expressed within various tissues of adult females, six of which are expressed within the fat body. All six Fox genes expressed in the fat body displayed dynamic expression profiles following a blood meal. We knocked down the 'fat body Foxes' through RNAi to determine whether these 'knockdowns' hindered amino acid-induced vitellogenin gene expression. We also determined the effect of these knockdowns on the number of eggs deposited following a blood meal. Knockdown of FoxN1, FoxN2, FoxL, and FoxO, had a negative effect on amino acid-induced vitellogenin gene expression and resulted in significantly fewer eggs laid. Our analysis stresses the importance of Fox transcription factors in regulating mosquito reproduction.
Figures

References
-
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. - PubMed
-
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. - PubMed
-
- Carlsson P, Mahlapuu M. Forkhead transcription factors: Key players in development and metabolism. Dev Biol. 2002;250:1–23. - PubMed
-
- Cheah PY, Chia W, Yang XH. Jumeaux, a novel Drosophila winged-helix family protein, is required for generating asymmetric sibling neuronal cell fates. Development. 2000;127:3325–3335. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

