Selective regulation of bone cell apoptosis by translational isoforms of the glucocorticoid receptor
- PMID: 17682054
- PMCID: PMC2168898
- DOI: 10.1128/MCB.00253-07
Selective regulation of bone cell apoptosis by translational isoforms of the glucocorticoid receptor
Abstract
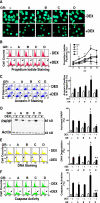
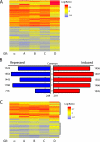
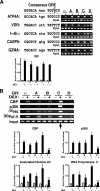
Glucocorticoids are widely used in the treatment of inflammatory and other diseases. However, high-dose or chronic administration often triggers troublesome side effects such as metabolic syndrome and osteoporosis. We recently described that one glucocorticoid receptor gene produces eight translational glucocorticoid receptor isoforms that have distinct gene-regulatory abilities. We show here that specific, but not all, glucocorticoid receptor isoforms induced apoptosis in human osteosarcoma U-2 OS bone cells. Whole human genome microarray analysis revealed that the majority of the glucocorticoid target genes were selectively regulated by specific glucocorticoid receptor isoforms. Real-time PCR experiments confirmed that proapoptotic enzymes necessary for cell death, granzyme A and caspase-6, were induced by specific glucocorticoid receptor isoforms. Chromatin immunoprecipitation assays further suggested that glucocorticoid receptor isoform-dependent induction of proapoptotic genes was likely due to selective coregulator recruitment and chromatin modification. Interestingly, the capabilities to transrepress proinflammatory genes were similar among glucocorticoid receptor isoforms. Together, these findings provide new evidence that translational glucocorticoid receptor isoforms can elicit distinct glucocorticoid responses and may be useful for the development of safe glucocorticoids with reduced side effects.
Figures







References
-
- Albiston, A. L., V. R. Obeyesekere, R. E. Smith, and Z. S. Krozowski. 1994. Cloning and tissue distribution of the human 11 beta-hydroxysteroid dehydrogenase type 2 enzyme. Mol. Cell. Endocrinol. 105:R11-R17. - PubMed
-
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25-29. - PMC - PubMed
-
- Bronnegard, M. 1996. Steroid receptor number. Individual variation and downregulation by treatment. Am. J. Respir. Crit. Care Med. 154:S28-S32, S32-S33. - PubMed
-
- Chrysis, D., F. Zaman, A. S. Chagin, M. Takigawa, and L. Savendahl. 2005. Dexamethasone induces apoptosis in proliferative chondrocytes through activation of caspases and suppression of the Akt-phosphatidylinositol 3′-kinase signaling pathway. Endocrinology 146:1391-1397. - PubMed
-
- Chua, C. C., B. H. Chua, Z. Chen, C. Landy, and R. C. Hamdy. 2003. Dexamethasone induces caspase activation in murine osteoblastic MC3T3-E1 cells. Biochim. Biophys. Acta 1642:79-85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
