Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation
- PMID: 17682057
- PMCID: PMC2168892
- DOI: 10.1128/MCB.00838-07
Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation
Abstract
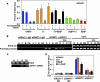
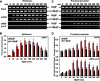
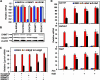
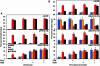
Transcriptional regulation of gene expression requires posttranslational modification of histone proteins, which, in concert with chromatin-remodeling factors, modulate chromatin structure. Exposure to environmental agents may interfere with specific histone modifications and derail normal patterns of gene expression. To test this hypothesis, we coexposed cells to binary mixtures of benzo[a]pyrene (B[a]P), an environmental procarcinogen that activates Cyp1a1 transcriptional responses mediated by the aryl hydrocarbon receptor (AHR), and chromium, a carcinogenic heavy metal that represses B[a]P-inducible AHR-mediated gene expression. We show that chromium cross-links histone deacetylase 1-DNA methyltransferase 1 (HDAC1-DNMT1) complexes to Cyp1a1 promoter chromatin and inhibits histone marks induced by AHR-mediated gene transactivation, including phosphorylation of histone H3 Ser-10, trimethylation of H3 Lys-4, and various acetylation marks in histones H3 and H4. These changes inhibit RNA polymerase II recruitment without affecting the kinetics of AHR DNA binding. HDAC1 and DNMT1 inhibitors or depletion of HDAC1 or DNMT1 with siRNAs blocks chromium-induced transcriptional repression by decreasing the interaction of these proteins with the Cyp1a1 promoter and allowing histone acetylation to proceed. By inhibiting Cyp1a1 expression, chromium stimulates the formation of B[a]P DNA adducts. Epigenetic modification of gene expression patterns may be a key element of the developmental and carcinogenic outcomes of exposure to chromium and to other environmental agents.
Figures




Similar articles
-
HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation.Biochim Biophys Acta. 2007 Sep-Oct;1769(9-10):569-78. doi: 10.1016/j.bbaexp.2007.07.002. Epub 2007 Jul 20. Biochim Biophys Acta. 2007. PMID: 17707923 Free PMC article.
-
Chromium inhibits transcription from polycyclic aromatic hydrocarbon-inducible promoters by blocking the release of histone deacetylase and preventing the binding of p300 to chromatin.J Biol Chem. 2004 Feb 6;279(6):4110-9. doi: 10.1074/jbc.M310800200. Epub 2003 Nov 18. J Biol Chem. 2004. PMID: 14625279
-
Butyrate alters expression of cytochrome P450 1A1 and metabolism of benzo[a]pyrene via its histone deacetylase activity in colon epithelial cell models.Arch Toxicol. 2017 May;91(5):2135-2150. doi: 10.1007/s00204-016-1887-4. Epub 2016 Nov 9. Arch Toxicol. 2017. PMID: 27830268
-
Chromatin modifications remodel cardiac gene expression.Cardiovasc Res. 2014 Jul 1;103(1):7-16. doi: 10.1093/cvr/cvu122. Epub 2014 May 8. Cardiovasc Res. 2014. PMID: 24812277 Review.
-
Readers, writers, and erasers: chromatin as the whiteboard of heart disease.Circ Res. 2015 Mar 27;116(7):1245-53. doi: 10.1161/CIRCRESAHA.116.303630. Circ Res. 2015. PMID: 25814685 Free PMC article. Review.
Cited by
-
Epigenetic alterations of CXCL5 in Cr(VI)-induced carcinogenesis.Sci Total Environ. 2022 Sep 10;838(Pt 1):155713. doi: 10.1016/j.scitotenv.2022.155713. Epub 2022 Jun 1. Sci Total Environ. 2022. PMID: 35660107 Free PMC article.
-
Hexavalent chromium increases the metabolism and genotoxicity of aromatic amine carcinogens 4-aminobiphenyl and β-naphthylamine in immortalized human lung epithelial cells.Toxicol Appl Pharmacol. 2022 Aug 15;449:116095. doi: 10.1016/j.taap.2022.116095. Epub 2022 Jun 2. Toxicol Appl Pharmacol. 2022. PMID: 35662664 Free PMC article.
-
Ah Receptor Pathway Intricacies; Signaling Through Diverse Protein Partners and DNA-Motifs.Toxicol Res (Camb). 2015 Sep 1;4(5):1143-1158. doi: 10.1039/c4tx00236a. Epub 2015 Mar 17. Toxicol Res (Camb). 2015. PMID: 26783425 Free PMC article.
-
Genetic and epigenetic modulations in toxicity: The two-sided roles of heavy metals and polycyclic aromatic hydrocarbons from the environment.Toxicol Rep. 2024 May 4;12:502-519. doi: 10.1016/j.toxrep.2024.04.010. eCollection 2024 Jun. Toxicol Rep. 2024. PMID: 38774476 Free PMC article. Review.
-
Nonadditive effects of PAHs on Early Vertebrate Development: mechanisms and implications for risk assessment.Toxicol Sci. 2008 Sep;105(1):5-23. doi: 10.1093/toxsci/kfm303. Epub 2007 Dec 20. Toxicol Sci. 2008. PMID: 18156145 Free PMC article.
References
-
- Alcedo, J. A., M. Misra, J. W. Hamilton, and K. E. Wetterhahn. 1994. The genotoxic carcinogen chromium(VI) alters the metal-inducible expression but not the basal expression of the metallothionein gene in vivo. Carcinogenesis 15:1089-1092. - PubMed
-
- Barchowsky, A., and K. A. O'Hara. 2003. Metal-induced cell signaling and gene activation in lung diseases. Free Radic. Biol. Med. 34:1130-1135. - PubMed
-
- Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315-326. - PubMed
-
- Brylawski, B. P., G. J. Tsongalis, M. Cordeiro-Stone, W. T. May, L. D. Comeau, and D. G. Kaufman. 1993. Association of putative origins of replication with the nuclear matrix in normal human fibroblasts. Cancer Res. 53:3865-3868. - PubMed
-
- Cefalu, W. T., Z. Q. Wang, X. H. Zhang, L. C. Baldor, and J. C. Russell. 2002. Oral chromium picolinate improves carbohydrate and lipid metabolism and enhances skeletal muscle Glut-4 translocation in obese, hyperinsulinemic (JCR-LA corpulent) rats. J. Nutr. 132:1107-1114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
