Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival
- PMID: 17682125
- PMCID: PMC2077319
- DOI: 10.1182/blood-2007-05-090142
Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival
Abstract
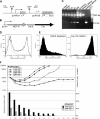
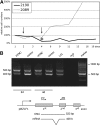
Many cells latently infected with Epstein-Barr virus (EBV), including certain virus-associated tumors, express latent membrane protein 2A (LMP2A), suggesting an important role for this protein in viral latency and oncogenesis. LMP2A mimics B-cell receptor signaling but can also act as a decoy receptor blocking B-cell receptor (BCR) activation. Studies of peripheral B cells have not resolved this apparent contradiction because LMP2A seems to be dispensable for EBV-induced transformation of these B cells in vitro. We show here that LMP2A is essential for growth transformation of germinal center B cells, which do not express the genuine BCR because of deleterious somatic hypermutations in their immunoglobulin genes. BCR-positive (BCR(+)) and BCR-negative (BCR(-)) B cells are readily transformed with a recombinant EBV encoding a conditional, floxed LMP2A allele, but the survival and continued proliferation of both BCR(+) and BCR(-) B cells is strictly dependent on LMP2A. These findings indicate that LMP2A has potent, distinct antiapoptotic and/or transforming characteristics and point to its role as an indispensable BCR mimic in certain B cells from which human B-cell tumors such as Hodgkin lymphoma originate.
Figures

References
-
- Vol 70. Lyon, France: IARC; 1997. Epstein-Barr virus and Kaposi's sarcoma herpesvirus/human herpesvirus 8: International Agency for Research on Cancer. Anonymous.
-
- Rickinson AB, Kieff E. Epstein-Barr virus. In: Knipe DM, Howley PM, Griffin DE, et al., editors. Fields Virology. 5th ed. Vol 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2655–2700.
-
- Kieff E, Rickinson AB. Epstein-Barr virus and its replication. In: Knipe DM, Howley PM, Griffin DE, et al., editors. Fields Virology. 5th ed. Vol 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2603–2654.
-
- Bell AI, Groves K, Kelly GL, et al. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J Gen Virol. 2006;87:2885–2890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

