Structure of the chloroplast ribosome: novel domains for translation regulation
- PMID: 17683199
- PMCID: PMC1939882
- DOI: 10.1371/journal.pbio.0050209
Structure of the chloroplast ribosome: novel domains for translation regulation
Abstract
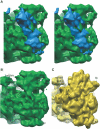
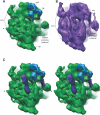
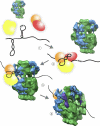
Gene expression in chloroplasts is controlled primarily through the regulation of translation. This regulation allows coordinate expression between the plastid and nuclear genomes, and is responsive to environmental conditions. Despite common ancestry with bacterial translation, chloroplast translation is more complex and involves positive regulatory mRNA elements and a host of requisite protein translation factors that do not have counterparts in bacteria. Previous proteomic analyses of the chloroplast ribosome identified a significant number of chloroplast-unique ribosomal proteins that expand upon a basic bacterial 70S-like composition. In this study, cryo-electron microscopy and single-particle reconstruction were used to calculate the structure of the chloroplast ribosome to a resolution of 15.5 A. Chloroplast-unique proteins are visualized as novel structural additions to a basic bacterial ribosome core. These structures are located at optimal positions on the chloroplast ribosome for interaction with mRNAs during translation initiation. Visualization of these chloroplast-unique structures on the ribosome, combined with mRNA cross-linking, allows us to propose a model for translation initiation in chloroplasts in which chloroplast-unique ribosomal proteins interact with plastid-specific translation factors and RNA elements to facilitate regulated translation of chloroplast mRNAs.
Conflict of interest statement
Figures




References
-
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19:149–168. - PubMed
-
- Watson JC, Surzycki SJ. Both the chloroplast and nuclear genome of Chlamydomonas reinhardtii share homology with Escherichia coli genes for transcriptional and translation components. Curr Genet. 1983;7:201–210. - PubMed
-
- Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R. Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine. 2005;23:1779–1783. - PubMed
-
- Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

