The circadian clock regulates auxin signaling and responses in Arabidopsis
- PMID: 17683202
- PMCID: PMC1939880
- DOI: 10.1371/journal.pbio.0050222
The circadian clock regulates auxin signaling and responses in Arabidopsis
Abstract
The circadian clock plays a pervasive role in the temporal regulation of plant physiology, environmental responsiveness, and development. In contrast, the phytohormone auxin plays a similarly far-reaching role in the spatial regulation of plant growth and development. Went and Thimann noted 70 years ago that plant sensitivity to auxin varied according to the time of day, an observation that they could not explain. Here we present work that explains this puzzle, demonstrating that the circadian clock regulates auxin signal transduction. Using genome-wide transcriptional profiling, we found many auxin-induced genes are under clock regulation. We verified that endogenous auxin signaling is clock regulated with a luciferase-based assay. Exogenous auxin has only modest effects on the plant clock, but the clock controls plant sensitivity to applied auxin. Notably, we found both transcriptional and growth responses to exogenous auxin are gated by the clock. Thus the circadian clock regulates some, and perhaps all, auxin responses. Consequently, many aspects of plant physiology not previously thought to be under circadian control may show time-of-day-specific sensitivity, with likely important consequences for plant growth and environmental responses.
Figures
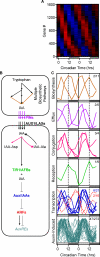
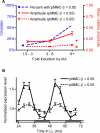


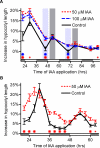
Comment in
-
Time to grow: circadian clock controls plant hormone signaling and response.PLoS Biol. 2007 Aug;5(8):e227. doi: 10.1371/journal.pbio.0050227. Epub 2007 Aug 7. PLoS Biol. 2007. PMID: 20076684 Free PMC article. No abstract available.
References
-
- Johnson CH, Kyriacou CP. Clock evolution and adaptation: Whence and whither? In: Hall AJW, McWatters HG, editors. Endogenous plant rhythms. Ames (Iowa): Blackwell Synergy; 2005. pp. 237–260.
-
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. - PubMed
-
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. - PubMed
-
- Salome PA, McClung CR. The Arabidopsis thaliana clock. J Biol Rhythms. 2004;19:425–435. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

