Length of guanosine homopolymeric repeats modulates promoter activity of subfamily II tpr genes of Treponema pallidum ssp. pallidum
- PMID: 17683506
- PMCID: PMC3006228
- DOI: 10.1111/j.1574-695X.2007.00303.x
Length of guanosine homopolymeric repeats modulates promoter activity of subfamily II tpr genes of Treponema pallidum ssp. pallidum
Abstract
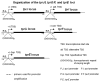
In Treponema pallidum, homopolymeric guanosine repeats of varying length are present upstream of both Subfamily I (tprC, D, F and I) and II (tprE, G and J) tpr genes, a group of potential virulence factors, immediately upstream of the +1 nucleotide. To investigate the influence of these poly-G sequences on promoter activity, tprE, G, J, F and I promoter regions containing homopolymeric tracts with different numbers of Gs, the ribosomal binding site and start codon were cloned in frame with the green fluorescent protein reporter gene (GFP), and promoter activity was measured both as fluorescence emission from Escherichia coli cultures transformed with the different plasmid constructs and using quantitative RT-PCR. For tprJ, G and E-derived clones, fluorescence was significantly higher with constructs containing eight Gs or fewer, while plasmids containing the same promoters with none or more Gs gave modest or no signal above the background. In contrast, tprF/I-derived clones induced similar levels of fluorescence regardless of the number of Gs within the promoter. GFP mRNA quantification showed that all of the promoters induced measurable transcription of the GFP gene; however, only for Subfamily II promoters was message synthesis inversely correlated to the number of Gs in the construct.
Figures




Similar articles
-
TP0262 is a modulator of promoter activity of tpr Subfamily II genes of Treponema pallidum ssp. pallidum.Mol Microbiol. 2009 Jun;72(5):1087-99. doi: 10.1111/j.1365-2958.2009.06712.x. Epub 2009 Apr 30. Mol Microbiol. 2009. PMID: 19432808 Free PMC article.
-
Gene organization and transcriptional analysis of the tprJ, tprI, tprG, and tprF loci in Treponema pallidum strains Nichols and Sea 81-4.J Bacteriol. 2005 Sep;187(17):6084-93. doi: 10.1128/JB.187.17.6084-6093.2005. J Bacteriol. 2005. PMID: 16109950 Free PMC article.
-
Treponema pallidum, the stealth pathogen, changes, but how?Mol Microbiol. 2009 Jun;72(5):1081-6. doi: 10.1111/j.1365-2958.2009.06711.x. Epub 2009 May 4. Mol Microbiol. 2009. PMID: 19432802 Free PMC article.
-
Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity.Microbes Infect. 2004 Jul;6(8):725-37. doi: 10.1016/j.micinf.2004.04.001. Microbes Infect. 2004. PMID: 15207819
-
Transcription of TP0126, Treponema pallidum putative OmpW homolog, is regulated by the length of a homopolymeric guanosine repeat.Infect Immun. 2015 Jun;83(6):2275-89. doi: 10.1128/IAI.00360-15. Epub 2015 Mar 23. Infect Immun. 2015. PMID: 25802057 Free PMC article.
Cited by
-
A novel pan-proteome array for high-throughput profiling of the humoral response to Treponema pallidum.iScience. 2024 Jul 31;27(9):110618. doi: 10.1016/j.isci.2024.110618. eCollection 2024 Sep 20. iScience. 2024. PMID: 39262771 Free PMC article.
-
TP0262 is a modulator of promoter activity of tpr Subfamily II genes of Treponema pallidum ssp. pallidum.Mol Microbiol. 2009 Jun;72(5):1087-99. doi: 10.1111/j.1365-2958.2009.06712.x. Epub 2009 Apr 30. Mol Microbiol. 2009. PMID: 19432808 Free PMC article.
-
Treponema pallidum, the syphilis spirochete: making a living as a stealth pathogen.Nat Rev Microbiol. 2016 Dec;14(12):744-759. doi: 10.1038/nrmicro.2016.141. Epub 2016 Oct 10. Nat Rev Microbiol. 2016. PMID: 27721440 Free PMC article. Review.
-
Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens.Nat Rev Microbiol. 2009 Jul;7(7):493-503. doi: 10.1038/nrmicro2145. Epub 2009 Jun 8. Nat Rev Microbiol. 2009. PMID: 19503065 Free PMC article. Review.
-
Identification of the Treponema pallidum subsp. pallidum TP0092 (RpoE) regulon and its implications for pathogen persistence in the host and syphilis pathogenesis.J Bacteriol. 2013 Feb;195(4):896-907. doi: 10.1128/JB.01973-12. Epub 2012 Dec 14. J Bacteriol. 2013. PMID: 23243302 Free PMC article.
References
-
- Arhin FF, Moreau F, Coulton JW, Mills EL. Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can J Microbiol. 1998;44:56–63. - PubMed
-
- Bayliss CD, Dixon KM, Moxon ER. Simple sequence repeats (microsatellites): mutational mechanisms and contributions to bacterial pathogenesis. A meeting review. FEMS Immunol Med Microbiol. 2004;40:11–19. - PubMed
-
- Centurion-Lara A, Castro C, Van Voorhis WC, Lukehart SA. Two 16S–23S ribosomal DNA intergenic regions in different Treponema pallidum subspecies contain tRNA genes. FEMS Microbiol Lett. 1996;143:235–240. - PubMed
-
- Dornberger U, Leijon M, Fritzsche H. High base pair opening rates in tracts of GC base pairs. J Biol Chem. 1999;274:6957–6962. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
