An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer
- PMID: 17683518
- PMCID: PMC2374988
- DOI: 10.1186/gb-2007-8-8-r157
An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer
Abstract
Background: Estrogen receptor (ER)-negative breast cancer specimens are predominantly of high grade, have frequent p53 mutations, and are broadly divided into HER2-positive and basal subtypes. Although ER-negative disease has overall worse prognosis than does ER-positive breast cancer, not all ER-negative breast cancer patients have poor clinical outcome. Reliable identification of ER-negative tumors that have a good prognosis is not yet possible.
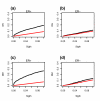
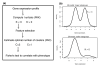
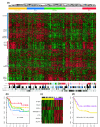
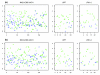
Results: We apply a recently proposed feature selection method in an integrative analysis of three major microarray expression datasets to identify molecular subclasses and prognostic markers in ER-negative breast cancer. We find a subclass of basal tumors, characterized by over-expression of immune response genes, which has a better prognosis than the rest of ER-negative breast cancers. Moreover, we show that, in contrast to ER-positive tumours, the majority of prognostic markers in ER-negative breast cancer are over-expressed in the good prognosis group and are associated with activation of complement and immune response pathways. Specifically, we identify an immune response related seven-gene module and show that downregulation of this module confers greater risk for distant metastasis (hazard ratio 2.02, 95% confidence interval 1.2-3.4; P = 0.009), independent of lymph node status and lymphocytic infiltration. Furthermore, we validate the immune response module using two additional independent datasets.
Conclusion: We show that ER-negative basal breast cancer is a heterogeneous disease with at least four main subtypes. Furthermore, we show that the heterogeneity in clinical outcome of ER-negative breast cancer is related to the variability in expression levels of complement and immune response pathway genes, independent of lymphocytic infiltration.
Figures

References
-
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

