Establishment and mitotic stability of an extra-chromosomal mammalian replicon
- PMID: 17683605
- PMCID: PMC1959191
- DOI: 10.1186/1471-2121-8-33
Establishment and mitotic stability of an extra-chromosomal mammalian replicon
Abstract
Background: Basic functions of the eukaryotic nucleus, like transcription and replication, are regulated in a hierarchic fashion. It is assumed that epigenetic factors influence the efficiency and precision of these processes. In order to uncouple local and long-range epigenetic features we used an extra-chromosomal replicon to study the requirements for replication and segregation and compared its behavior to that of its integrated counterpart.
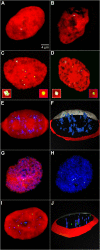
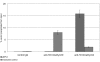
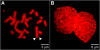
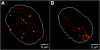
Results: The autonomous replicon replicates in all eukaryotic cells and is stably maintained in the absence of selection but, as other extra-chromosomal replicons, its establishment is very inefficient. We now show that following establishment the vector is stably associated with nuclear compartments involved in gene expression and chromosomal domains that replicate at the onset of S-phase. While the vector stays autonomous, its association with these compartments ensures the efficiency of replication and mitotic segregation in proliferating cells.
Conclusion: Using this novel minimal model system we demonstrate that relevant functions of the eukaryotic nucleus are strongly influenced by higher nuclear architecture. Furthermore our findings have relevance for the rational design of episomal vectors to be used for genetic modification of cells: in order to improve such constructs with respect to efficiency elements have to be identified which ensure that such constructs reach regions of the nucleus favorable for replication and transcription.
Figures


Similar articles
-
Alphavirus cDNA-based expression vectors: effects of RNA transcription and nuclear export.Biotechnol Bioeng. 2003 Mar 5;81(5):553-62. doi: 10.1002/bit.10496. Biotechnol Bioeng. 2003. PMID: 12514804
-
Cell cycle dependent histone dynamics of an episomal non-viral vector.Gene. 2009 Jun 15;439(1-2):95-101. doi: 10.1016/j.gene.2009.03.010. Epub 2009 Mar 21. Gene. 2009. PMID: 19306916
-
Designing nonviral vectors for efficient gene transfer and long-term gene expression.Mol Ther. 2006 Nov;14(5):613-26. doi: 10.1016/j.ymthe.2006.03.026. Epub 2006 Jun 19. Mol Ther. 2006. PMID: 16784894 Review.
-
Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix.Nat Cell Biol. 2000 Mar;2(3):182-4. doi: 10.1038/35004061. Nat Cell Biol. 2000. PMID: 10707091 No abstract available.
-
Initiation of replication in mammalian chromosomes.Crit Rev Eukaryot Gene Expr. 1992;2(4):359-81. Crit Rev Eukaryot Gene Expr. 1992. PMID: 1486243 Review.
Cited by
-
Genome-wide profiling of S/MAR-based replicon contact sites.Nucleic Acids Res. 2017 Jul 27;45(13):7841-7854. doi: 10.1093/nar/gkx522. Nucleic Acids Res. 2017. PMID: 28609784 Free PMC article.
-
The Herpes Simplex Virus 1 Protein ICP4 Acts as both an Activator and a Repressor of Host Genome Transcription during Infection.Mol Cell Biol. 2021 Sep 24;41(10):e0017121. doi: 10.1128/MCB.00171-21. Epub 2021 Jul 12. Mol Cell Biol. 2021. PMID: 34251885 Free PMC article.
-
pEPito: a significantly improved non-viral episomal expression vector for mammalian cells.BMC Biotechnol. 2010 Mar 15;10:20. doi: 10.1186/1472-6750-10-20. BMC Biotechnol. 2010. PMID: 20230618 Free PMC article.
-
Genetic modification of cancer cells using non-viral, episomal S/MAR vectors for in vivo tumour modelling.PLoS One. 2012;7(10):e47920. doi: 10.1371/journal.pone.0047920. Epub 2012 Oct 26. PLoS One. 2012. PMID: 23110132 Free PMC article.
-
Efficient episomal gene transfer to human hepatic cells using the pFAR4-S/MAR vector.Mol Biol Rep. 2019 Jun;46(3):3203-3211. doi: 10.1007/s11033-019-04777-9. Epub 2019 Apr 12. Mol Biol Rep. 2019. PMID: 30980265
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

