Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae?
- PMID: 17683611
- PMCID: PMC2045110
- DOI: 10.1186/1471-2148-7-132
Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae?
Abstract
Background: Molecular lock-and-key systems are common among reproductive proteins, yet their evolution remains a major puzzle in evolutionary biology. In the Brassicaceae, the genes encoding self-incompatibility have been identified, but technical challenges currently prevent detailed analyses of the molecular interaction between the male and female components. In the present study, we investigate sequence polymorphism in the female specificity determinant SRK of Arabidopsis halleri from throughout Europe. Using a comparative approach based on published SRK sequences in A. lyrata and Brassica, we track the signature of frequency-dependent selection acting on these genes at the codon level. Using simulations, we evaluate power and accuracy of our approach and estimate the proportion of codon sites involved in the molecular interaction.
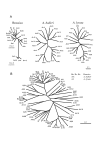
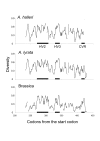
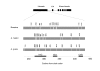
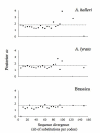
Results: We identified several members of the S-gene family, together with 22 putative S-haplotypes. Linkage to the S-locus and the presence of a kinase domain were formally demonstrated for four and six of these haplotypes, respectively, and sequence polymorphism was extremely high. Twenty-five codons showed signs of positive selection in at least one species, and clustered significantly (but not exclusively) within hypervariable regions. We checked that this clustering was not an artifact due to variation in evolution rate at synonymous sites. Simulations revealed that the analysis was highly accurate, thus providing a reliable set of candidates for future functional analyses, but with an overall power not higher than 60 %. Assuming similar power, we infer from our results that about 23% of all codons in the S-domain may actually be involved in recognition. Interestingly, while simulations demonstrated that this comparison remained reliable even at very high levels of divergence, codons identified in Brassica had higher posterior rates of non-synonymous to synonymous substitutions than codons identified in A. halleri or A. lyrata, possibly suggesting more intense selection in Brassica.
Conclusion: The signature of balancing selection can be identified reliably at the codon level even in cases of very high sequence divergence, provided that a sufficiently large set of sequences are analyzed. Altogether, our results indicate that a large proportion of codons may be involved in recognition and confirm the particular importance of hypervariable regions. The more intense signature of positive selection detected in Brassica suggests that allelic diversification in this genus was very recent, possibly following a recent demographic bottleneck.
Figures
Similar articles
-
The evolution and diversification of S-locus haplotypes in the Brassicaceae family.Genetics. 2009 Mar;181(3):977-84. doi: 10.1534/genetics.108.090837. Epub 2008 Dec 15. Genetics. 2009. PMID: 19087966 Free PMC article.
-
The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr.Mol Biol Evol. 2006 Sep;23(9):1741-50. doi: 10.1093/molbev/msl042. Epub 2006 Jun 16. Mol Biol Evol. 2006. PMID: 16782760
-
Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata.Mol Biol Evol. 2003 Nov;20(11):1741-53. doi: 10.1093/molbev/msg170. Epub 2003 Jun 27. Mol Biol Evol. 2003. PMID: 12832651
-
Molecular evolution of the s locus controlling mating in the brassicaceae.Plant Biol (Stuttg). 2004 Mar-Apr;6(2):109-18. doi: 10.1055/s-2004-817804. Plant Biol (Stuttg). 2004. PMID: 15045661 Review.
-
Molecular aspects of self-incompatibility in Brassica species.Plant Cell Physiol. 2001 Jun;42(6):560-5. doi: 10.1093/pcp/pce075. Plant Cell Physiol. 2001. PMID: 11427674 Review.
Cited by
-
Sporophytic self-incompatibility genes and mating system variation in Arabis alpina.Ann Bot. 2011 Sep;108(4):699-713. doi: 10.1093/aob/mcr157. Epub 2011 Aug 5. Ann Bot. 2011. PMID: 21821832 Free PMC article.
-
MIK2 is a candidate gene of the S-locus for sporophytic self-incompatibility in chicory (Cichorium intybus, Asteraceae).Front Plant Sci. 2023 Jun 2;14:1204538. doi: 10.3389/fpls.2023.1204538. eCollection 2023. Front Plant Sci. 2023. PMID: 37332702 Free PMC article.
-
Transition to Self-compatibility Associated With Dominant S-allele in a Diploid Siberian Progenitor of Allotetraploid Arabidopsis kamchatica Revealed by Arabidopsis lyrata Genomes.Mol Biol Evol. 2023 Jul 5;40(7):msad122. doi: 10.1093/molbev/msad122. Mol Biol Evol. 2023. PMID: 37432770 Free PMC article.
-
Evolutionary patterns at the RNase based gametophytic self - incompatibility system in two divergent Rosaceae groups (Maloideae and Prunus).BMC Evol Biol. 2010 Jun 28;10:200. doi: 10.1186/1471-2148-10-200. BMC Evol Biol. 2010. PMID: 20584298 Free PMC article.
-
Repeated adaptive introgression at a gene under multiallelic balancing selection.PLoS Genet. 2008 Aug 29;4(8):e1000168. doi: 10.1371/journal.pgen.1000168. PLoS Genet. 2008. PMID: 18769722 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

